Difference between revisions of "NumMethodsPDEs"
| (208 intermediate revisions by the same user not shown) | |||
| Line 4: | Line 4: | ||
=Introduction= | =Introduction= | ||
| + | ==Theory== | ||
| − | + | In this tutorial we're going to look at how to solve Partial Differential Equations (PDEs) ''numerically''. That is, on a computer, as opposed to analytically, as we would do using pencil and paper. PDEs crop up a good deal when we describe things in the natural world--e.g. heat flow, or the distribution of electrical charge--and so methods for their solution prove to be widely useful when writing scientific models. | |
| − | + | Given the relatively large amount of material that we're about to cover, let's list the main take-home messages straight away: | |
| + | * Finding a numerical solution to a PDE often boils down (after discritisation) to solving a system of linear equations, of the (matrix) form '''Ax'''='''b'''. | ||
| + | * Third-party linear algebra libraries will be much faster and less error prone than your own code for this task. Many of these libraries will also run in parallel. | ||
| + | * By writing code at and above the level of a call to a linear algebra library, we can focus on the higher-level task of doing the science. | ||
| + | * If the matrix '''A''' is sparse then iterative methods will be ''much'' faster than performing a direct method for solution (such as an LU decomposition). | ||
| − | + | ==Practice== | |
| + | In this tutorial we'll take a look at a number of example programs which solve equations using linear algebra packages. These packages include LAPACK, PETSc, PLASMA and MAGMA. The examples will be based upon the solution of a PDE using the Finite Difference Method (FDM), although we'll also touch upon the theory for an alternative approach--the Finite Element Method (FEM). | ||
| + | To download the code for the examples, type: | ||
| + | <pre> | ||
| + | svn co https://svn.ggy.bris.ac.uk/subversion-open/num-methods1 ./num-methods1 | ||
| + | </pre> | ||
| − | <math> f(x + h) | + | LAPACK (which stands for Linear Algebra PACKage), is the oldest of the packages considered. It has a venerable history and is still highly relevant for some problems today. The project website--which contains download instructions and a great deal of useful documentation--is: http://www.netlib.org/lapack. LAPACK is built on top of the Basic Linear Algebra Subprograms (BLAS) (http://www.netlib.org/blas), but provides a higher level functionality. |
| + | |||
| + | Before looking at our first example, we'll start building a 'reference' implementation of LAPACK, as it will take several minutes. First change directory to the location of the LAPACK code that has been bundled into the repository of examples: | ||
| + | |||
| + | <pre> | ||
| + | cd num-methods1/lapack-3.3.1 | ||
| + | </pre> | ||
| + | |||
| + | and build the libraries, by typing: | ||
| + | |||
| + | <pre> | ||
| + | make lib | ||
| + | </pre> | ||
| + | |||
| + | =Theory: Looking at Nature (& a quick philosophical aside)= | ||
| + | |||
| + | When we observe nature, certain patterns crop up again and again. For example, in the the field of electrostatics, if a function <math>f</math> describes a distribution of electric charge, then Poisson's equation: | ||
| + | |||
| + | :<math>{\nabla}^2 u = f.</math> | ||
| + | |||
| + | gives the electric potential <math>u</math>. | ||
| + | |||
| + | However, Poisson's equation also describes the steady state temperature of a material when subjected to some heating and also crops up when considering gravitational potentials.. | ||
| + | |||
| + | What's going on here? Why are we using the same equation? Is Poisson's equation fundamental in some way? Well, yes I suppose it is. However, we can look at it another way and say that Poisson's equation is the way we describe--using the language of maths--steady state phenomena which involve potentials. OK, this sounds a bit out there, but consider the following very general equation for a moment (it's a second-order linear equation in two variables): | ||
| + | |||
| + | <math>Au_{xx} + Bu_{xy} + Cu_{yy} + Du_x + Eu_y + Fu = G</math> | ||
| + | |||
| + | It turns out that we can categorise certain instances of this equation and then relate these categories to the kind of phenomena that they describe: | ||
| + | |||
| + | * '''Parabolic''': <math>B^2 - 4AC = 0</math>. This family of equations describe heat flow and diffusion processes. | ||
| + | * '''Hyperbolic''': <math>B^2 - 4AC > 0</math>. Describe vibrating systems and wave motion. | ||
| + | * '''Elliptic''': <math>B^2 - 4AC < 0</math>. Steady-state phenomena. | ||
| + | |||
| + | That's pretty handy! OK, we've had our philosophical aside, on with the show. | ||
| + | |||
| + | =Practice: Guassian Elimination= | ||
| + | |||
| + | While the libraries are building, consider the following system of linear equations: | ||
| + | |||
| + | :<math>\begin{alignat}{7} | ||
| + | x &&\; + \;&& 3y &&\; - \;&& 2z &&\; = \;&& 5 & \\ | ||
| + | 3x &&\; + \;&& 5y &&\; + \;&& 6z &&\; = \;&& 7 & \\ | ||
| + | 2x &&\; + \;&& 4y &&\; + \;&& 3z &&\; = \;&& 8 & | ||
| + | \end{alignat}</math> | ||
| + | |||
| + | (From http://en.wikipedia.org/wiki/System_of_linear_equations.) | ||
| + | |||
| + | which we can write in matrix form: | ||
| + | |||
| + | :<math> | ||
| + | \begin{bmatrix}1 & 3 & -2\\3 & 5 & 6\\2 & 4 & 3\end{bmatrix} | ||
| + | \begin{bmatrix}x\\y\\z\end{bmatrix} | ||
| + | = | ||
| + | \begin{bmatrix}5\\7\\8\end{bmatrix} | ||
| + | </math> | ||
| + | |||
| + | The following computation shows Gauss-Jordan elimination applied a further simplified (assumed the x, y and z) matrix version ('''Ax'''='''b''') of the above system, where the operations include: | ||
| + | # top row x3, subtract from 2nd row | ||
| + | # top row x2, subtract from 3rd row | ||
| + | # dived the 2nd row by 4 | ||
| + | # multiply the 2nd row by 2, add to the 3rd row | ||
| + | etc: | ||
| + | |||
| + | :<math>\left[\begin{array}{rrr|r} | ||
| + | 1 & 3 & -2 & 5 \\ | ||
| + | 3 & 5 & 6 & 7 \\ | ||
| + | 2 & 4 & 3 & 8 | ||
| + | \end{array}\right]</math><math>\sim | ||
| + | \left[\begin{array}{rrr|r} | ||
| + | 1 & 3 & -2 & 5 \\ | ||
| + | 0 & -4 & 12 & -8 \\ | ||
| + | 2 & 4 & 3 & 8 | ||
| + | \end{array}\right]</math><math>\sim | ||
| + | \left[\begin{array}{rrr|r} | ||
| + | 1 & 3 & -2 & 5 \\ | ||
| + | 0 & -4 & 12 & -8 \\ | ||
| + | 0 & -2 & 7 & -2 | ||
| + | \end{array}\right]</math><math>\sim | ||
| + | \left[\begin{array}{rrr|r} | ||
| + | 1 & 3 & -2 & 5 \\ | ||
| + | 0 & 1 & -3 & 2 \\ | ||
| + | 0 & -2 & 7 & -2 | ||
| + | \end{array}\right]</math><math>\sim | ||
| + | \left[\begin{array}{rrr|r} | ||
| + | 1 & 3 & -2 & 5 \\ | ||
| + | 0 & 1 & -3 & 2 \\ | ||
| + | 0 & 0 & 1 & 2 | ||
| + | \end{array}\right]</math><math>\sim | ||
| + | \left[\begin{array}{rrr|r} | ||
| + | 1 & 3 & -2 & 5 \\ | ||
| + | 0 & 1 & 0 & 8 \\ | ||
| + | 0 & 0 & 1 & 2 | ||
| + | \end{array}\right]</math><math>\sim | ||
| + | \left[\begin{array}{rrr|r} | ||
| + | 1 & 3 & 0 & 9 \\ | ||
| + | 0 & 1 & 0 & 8 \\ | ||
| + | 0 & 0 & 1 & 2 | ||
| + | \end{array}\right]</math><math>\sim | ||
| + | \left[\begin{array}{rrr|r} | ||
| + | 1 & 0 & 0 & -15 \\ | ||
| + | 0 & 1 & 0 & 8 \\ | ||
| + | 0 & 0 & 1 & 2 | ||
| + | \end{array}\right].</math> | ||
| + | |||
| + | OK, so far so good? Let's try our first code example: | ||
| + | |||
| + | <pre> | ||
| + | cd ~/num-methods1/examples/example1 | ||
| + | </pre> | ||
| + | |||
| + | To see how to solve the above matrix example using LAPACK take a look inside '''dgesv-example.f90'''. First the declarations: | ||
| + | |||
| + | <source lang="fortran"> | ||
| + | integer, parameter :: N = 3 | ||
| + | integer, parameter :: LDA = N ! leading dimension of A | ||
| + | integer, parameter :: LDB = N ! leading dimension of B | ||
| + | integer, parameter :: NRHS = 1 ! no. of RHS, i.e columns in b | ||
| + | integer, dimension(N) :: IPIV | ||
| + | integer :: INFO | ||
| + | integer :: i | ||
| + | real(kind=8), dimension(LDA,N) :: A ! LDAxN matrix | ||
| + | real(kind=8), dimension(LDB,NRHS) :: B ! LDBxNRHS matrix | ||
| + | </source> | ||
| + | |||
| + | Where, we see that the ''leading dimension'' of a matrix is used to specify the number of rows of storage. Note also, that we could simultaneously solve for multiple vectors B, by specifying NRHS > 1. | ||
| + | |||
| + | Next, we set up the matrix A: | ||
| + | <source lang="fortran"> | ||
| + | ! ( 1 3 -2) | ||
| + | ! ( 3 5 6) | ||
| + | ! ( 2 4 3) | ||
| + | A(1,1) = 1.0d+0 | ||
| + | A(1,2) = 3.0d+0 | ||
| + | A(1,3) = -2.0d+0 | ||
| + | A(2,1) = 3.0d+0 | ||
| + | A(2,2) = 5.0d+0 | ||
| + | A(2,3) = 6.0d+0 | ||
| + | A(3,1) = 2.0d+0 | ||
| + | A(3,2) = 4.0d+0 | ||
| + | A(3,3) = 3.0d+0 | ||
| + | </source> | ||
| + | |||
| + | And the vector B: | ||
| + | <source lang="fortran"> | ||
| + | ! ( 5) | ||
| + | ! ( 7) | ||
| + | ! ( 8) | ||
| + | B(1,1) = 5.0d+0 | ||
| + | B(2,1) = 7.0d+0 | ||
| + | B(3,1) = 8.0d+0 | ||
| + | </source> | ||
| + | |||
| + | Once we have all that in place, all we need to do is call the solver routine: | ||
| + | |||
| + | <source lang="fortran"> | ||
| + | call dgesv(N, NRHS, A, LDA, IPIV, B, LDB, INFO) | ||
| + | </source> | ||
| + | |||
| + | Note that the solution vector ''replaces'' the values in B. (The contents of A will be changed in place too, but more of that later.) | ||
| + | |||
| + | <source lang="fortran"> | ||
| + | ! Print the result vector X. | ||
| + | ! It should be: | ||
| + | ! (-15) | ||
| + | ! ( 8) | ||
| + | ! ( 2) | ||
| + | do ii=1,LDB | ||
| + | write(*,*) B(ii,1) | ||
| + | end do | ||
| + | </source> | ||
| + | |||
| + | Run the program to satisfy yourself that it works: | ||
| + | |||
| + | <pre> | ||
| + | cd num-methods1/examples/example1 | ||
| + | make | ||
| + | ./dgesv-example.exe | ||
| + | </pre> | ||
| + | |||
| + | =The Finite Difference Method= | ||
| + | |||
| + | In this section we'll look at solving PDEs using the Finite Difference Method. Finite difference equations have a long and illustrious past. Indeed, they were used by Newton-and-the-gang as a logical route in to talk about differential calculus. So, in fact, we're just going to work back in the other direction.. | ||
| + | |||
| + | ==Theory: Discritisation== | ||
| + | |||
| + | The functions and variables that we see in, for instance, Poisson's equation are continuous; meaning that they could have an infinite number of values. This is fine when solving equations analytically, but we are limited to considering only a '''finite''' number of values when solving PDEs using a digital computer. We need a way to discritise our problems. Happily, help is at hand from the (sometimes tortuous) Taylor series: We can approximate the value of a function at a point distant from x (i.e. displaced by an amount h) using a linear summation of derivatives of the function at x--a Taylor expansion: | ||
| + | |||
| + | :<math>f(x+h) \approx f(x) + \frac{f'(x)}{1!}h + \frac{f''(x)}{2!}h^2 + \cdots</math> | ||
| + | |||
| + | We can truncate this series after the first differential and rearrange, to get ourselves the '''forward-difference''' approximation: | ||
| + | |||
| + | :<math>f'(x) \approx \frac{f(x+h) - f(x)}{h}</math> | ||
| + | |||
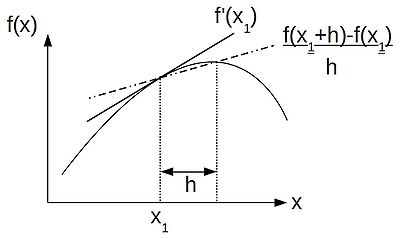
| + | Thus, following the finite difference approach, we can discritise our equations by replacing our differential operators by their difference approximations, as helpfully provided by a truncated Taylor series. If, like me, you like thinking visually, the schematic below shows the relationship between a differential operator--the gradient at a point <math>x_1</math> and the finite difference approximation to the gradient: | ||
| + | |||
| + | [[Image:forward-difference.jpg|400px|centre|thumbnail|A graphical representation of a forward-difference approximation to the gradient at a point <math>x_1</math>. The approximation gives way to the true value of <math>f'(x_1)</math> as the distance h shrinks to become infinitesimally small.]] | ||
| + | |||
| + | If we were to consider a displacement of -h instead, we would get the '''backward-difference''' approximation to <math>f'(x)</math>: | ||
| + | |||
| + | :<math>f'(x) \approx \frac{f(x) - f(x-h)}{h} </math>, | ||
| + | |||
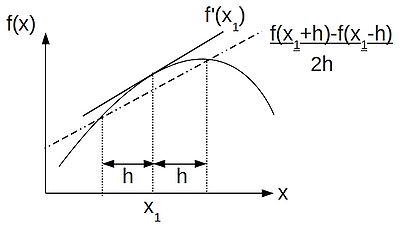
| + | Subtracting the backward difference from the forward difference we get the '''central-difference''' approximation: | ||
| + | |||
| + | :<math>f'(x) \approx \frac{1}{2h}(f(x+h) - f(x-h))</math>. | ||
| + | |||
| + | [[Image:central-difference.jpg|400px|centre|thumbnail|A graphical representation of a central-difference approximation to the gradient at a point <math>x_1</math>. The approximation gives way to the true value of <math>f'(x_1)</math> as the distance h shrinks to become infinitesimally small.]] | ||
| + | |||
| + | By retaining an extra term from the Taylor series, and some similar rearrangements, we can also formulate a finite difference approximation to the '''2nd differential'''. For example the central-difference formula for this is: | ||
| + | |||
| + | :<math>f''(x) \approx \frac{1}{h^2} [ f(x+h) - 2f(x) + f(x-h) ] </math>. | ||
| + | |||
| + | In a similar way we can derive formulae for approximations to partial derivatives. Armed with all these formulae, we can convert differential equations into difference equations, which we can solve on a computer. However, since we are dealing with approximations to the original equation(s), we must be vigilant that errors in our input and intermediate calculations do not accumulate such that our final solution is meaningless. | ||
| + | |||
| + | ==Theory: Eliptical Equations & the Discrete Laplacian Operator== | ||
| + | |||
| + | Now that we have our difference formulae, let's apply them to Laplace's equation in 2-dimensions: | ||
| + | |||
| + | :<math>\frac{\partial^2u}{\partial x^2} + \frac{\partial^2u}{\partial y^2} = 0</math> | ||
| + | |||
| + | This describes purely diffusive phenomena and can give us, for example, the steady state temperature over a 2D plate. Let's suppose that three sides of the plate are held at 100°C, while the fourth is held at 0°C. We can divide the plate into a number of grid cells: | ||
| + | |||
| + | [[Image:4x4-2D-laplacian-grid.jpg|300px|centre|thumbnail|A gridded representation of a plate with 3 sides kept at 100°C, 1 side at 0°C]] | ||
| + | |||
| + | Using the central difference approximation to a 2nd differential that we derived in the last section, we can rewrite the differential equation as a difference equation: | ||
| + | |||
| + | :<math>\frac{\partial^2u}{\partial x^2} + \frac{\partial^2u}{\partial y^2} \approx \frac{1}{h^2}(u^j_{i+1} -2u^j_i +u^j_{i-1}) + \frac{1}{k^2}(u^{j+1}_i -2u^j_i +u^{j-1}_i)</math> | ||
| + | |||
| + | where h is the grid spacing in the i-direction and k is the spacing the j-direction. If we set <math>h=k</math>, then: | ||
| + | |||
| + | :<math>u^j_{i+1} + u^j_{i-1} + u^{j+1}_i + u^{j-1}_i - 4u^j_i = 0</math> | ||
| + | |||
| + | solving for <math>u^j_i</math>: | ||
| + | |||
| + | :<math>u^j_i = \frac{1}{4}(u^j_{i+1} + u^j_{i-1} + u^{j+1}_i + u^{j-1}_i)</math> | ||
| + | |||
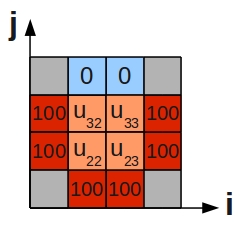
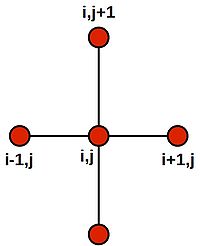
| + | Note that this last equation says--quite intuitively--that the value at the central grid point is equal to the average of its four neighbours. We can draw a picture of the '''stencil''' needed to compute the central value: | ||
| + | |||
| + | [[Image:five-point-stencil.jpg|200px|thumbnail|centre|Stencil for a 2D finite difference approximation to the 2nd differential]] | ||
| + | |||
| + | Centring the stencil on each of the 4 grid cells of the inner region (orange) in turn, we generate the following 4 difference equations: | ||
| + | |||
| + | :<math> | ||
| + | \begin{alignat}{5} | ||
| + | -4u_{22} & + u_{32} & + u_{23} & + 100 & + 100 & = 0 \\ | ||
| + | -4u_{32} & + 0 & + u_{33} & + u_{22} & + 100 & = 0 \\ | ||
| + | -4u_{23} & + u_{33} & + 100 & + 100 & + u_{22} & = 0 \\ | ||
| + | -4u_{33} & + 0 & + 100 & + u_{23} & + u_{32} & = 0 \\ | ||
| + | \end{alignat} | ||
| + | </math> | ||
| + | |||
| + | |||
| + | This system of equations can be written in matrix form: | ||
| + | :<math> | ||
| + | \begin{bmatrix}-4 & 1 & 1 & 0\\1 & -4 & 0 & 1\\1 & 0 & -4 & 1\\0 & 1 & 1 & -4\end{bmatrix} | ||
| + | \begin{bmatrix}u_{22}\\u_{32}\\u_{23}\\u_{33}\end{bmatrix} | ||
| + | = | ||
| + | \begin{bmatrix}-200\\-100\\-200\\-100\end{bmatrix} | ||
| + | </math> | ||
| + | |||
| + | Which may be summarised as the vector equation, '''Ax'''='''b''', which can be efficiently & conveniently solved using a library linear algebra routines, such as LAPACK, PETSc or PLASMA. | ||
| + | |||
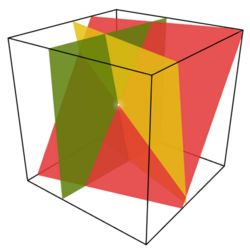
| + | [[Image:Secretsharing-3-point.png|250px|thumbnail|centre|The solution to a system of linear equations amounts to finding the intersection of lines, or planes, depending upon the number of unknown variables.]] | ||
| + | |||
| + | ==Practice: Solving Eliptical Problems using LAPACK== | ||
| + | |||
| + | Next, take a look at '''simple-laplace.f90'''. This program solves the 2D laplace equation for a 4x4 domain, as described above. This program only differs from the previous one in way in which we initialise the matrix '''A''' and vector '''b'''. To run the program type: | ||
| + | |||
| + | <pre> | ||
| + | ./simple-laplace.exe | ||
| + | </pre> | ||
| + | |||
| + | '''mxm-laplace.f90''' also solves our laplacian example, except that this time we can vary the size of the matrix and corresponding arrays for an mxm square domain: | ||
| + | |||
| + | <pre> | ||
| + | ./mxm-laplace.exe | ||
| + | </pre> | ||
| + | |||
| + | LAPACK also gives us access to a routine--'''dgesvx'''--which solves the same system of equations, but also gives us some error analysis. The '''x''' stands for e'''x'''pert. | ||
| + | |||
| + | <pre> | ||
| + | ./mxm-laplace-expert.exe | ||
| + | </pre> | ||
| + | |||
| + | ---- | ||
| + | |||
| + | '''Exercises''' | ||
| + | |||
| + | * Play around with '''mxm-laplace.f90'''. Try different domain sizes, and different boundary conditions. | ||
| + | |||
| + | ==Practice: Details of the LU Decomposition== | ||
| + | |||
| + | Actually, the LAPACK routine '''dgesv''' does not perform Gaussian elimination, but rather computes an '''LU decomposition''' and then solves the equation using two substitution steps--one forward and one backward. This is because for the same computational cost, we can factor '''A''' without knowing about '''b'''. We can then find '''x''' cheaply, for many different '''b'''. This is useful when solving parabolic equations, where '''x''' at time-step <math>j</math> forms the basis for '''b''' at time-step <math>j+1</math>. | ||
| + | |||
| + | OK, how to we obtain an LU decomposition? | ||
| + | |||
| + | The Guassian elimination process above can be expressed as a matrix multiplication. The elimination in the first column can be achieved by left multiplying '''A''' with a unit (1's on the diagonal) lower triangular matrix, <math>L_1</math>, with non-zeros only in the first column (this case is called an ''atomic unit lower triangular'' matrix), such as that schematically shown below: | ||
| + | |||
| + | :<math> | ||
| + | \begin{bmatrix} | ||
| + | 1 & 0 & 0 \\ | ||
| + | x & 1 & 0 \\ | ||
| + | y & 0 & 1 | ||
| + | \end{bmatrix} | ||
| + | </math> | ||
| + | |||
| + | Following our example, <math>L_1 A=A'</math> becomes: | ||
| + | |||
| + | :<math> | ||
| + | \begin{bmatrix} | ||
| + | 1 & 0 & 0 \\ | ||
| + | -3 & 1 & 0 \\ | ||
| + | -2 & 0 & 1 | ||
| + | \end{bmatrix} | ||
| + | \times | ||
| + | \begin{bmatrix} | ||
| + | 1 & 3 & -2 \\ | ||
| + | 3 & 5 & 6 \\ | ||
| + | 2 & 4 & 3 | ||
| + | \end{bmatrix} | ||
| + | = | ||
| + | \begin{bmatrix} | ||
| + | 1 & 3 & -2 \\ | ||
| + | 0 & -4 & 12 \\ | ||
| + | 0 & -2 & 7 | ||
| + | \end{bmatrix} | ||
| + | </math> | ||
| + | |||
| + | The eliminations in the second column are achieved by left multiplying <math>A'=L_1 A</math> by <math>L_2</math> (which is atomic unit lower triangular with non-zeros only in the second column): | ||
| + | |||
| + | :<math> | ||
| + | \begin{bmatrix} | ||
| + | 1 & 0 & 0 \\ | ||
| + | 0 & 1 & 0 \\ | ||
| + | 0 & -\frac{1}{2} & 1 | ||
| + | \end{bmatrix} | ||
| + | \times | ||
| + | \begin{bmatrix} | ||
| + | 1 & 0 & 0 \\ | ||
| + | -3 & 1 & 0 \\ | ||
| + | -2 & 0 & 1 | ||
| + | \end{bmatrix} | ||
| + | \times | ||
| + | \begin{bmatrix} | ||
| + | 1 & 3 & -2 \\ | ||
| + | 3 & 5 & 6 \\ | ||
| + | 2 & 4 & 3 | ||
| + | \end{bmatrix} | ||
| + | = | ||
| + | \begin{bmatrix} | ||
| + | 1 & 3 & -2 \\ | ||
| + | 0 & -4 & 12 \\ | ||
| + | 0 & 0 & 1 | ||
| + | \end{bmatrix} | ||
| + | </math> | ||
| + | |||
| + | We note that the result <math>L_2 L_1 A = A'' = U</math> is upper triangular. Rearranging we get, <math>A=L_1^{-1} \cdots L^{-1}_{n-2} L^{-1}_{n-1} U</math>. The inverse matrices, <math>L^{-1}</math>, are very easy to find as the inverse of an atomic unit lower triangular matrix is the same as the original save only for a change of sign in the off diagonal elements. The product is, again, unit lower triangular. Thus <math>A=LU</math>: | ||
| + | |||
| + | :<math> | ||
| + | \begin{bmatrix} | ||
| + | 1 & 3 & -2 \\ | ||
| + | 3 & 5 & 6 \\ | ||
| + | 2 & 4 & 3 | ||
| + | \end{bmatrix} | ||
| + | = | ||
| + | \begin{bmatrix} | ||
| + | 1 & 0 & 0 \\ | ||
| + | 0 & 1 & 0 \\ | ||
| + | 0 & \frac{1}{2} & 1 | ||
| + | \end{bmatrix} | ||
| + | \times | ||
| + | \begin{bmatrix} | ||
| + | 1 & 0 & 0 \\ | ||
| + | 3 & 1 & 0 \\ | ||
| + | 2 & 0 & 1 | ||
| + | \end{bmatrix} | ||
| + | \times | ||
| + | \begin{bmatrix} | ||
| + | 1 & 3 & -2 \\ | ||
| + | 0 & -4 & 12 \\ | ||
| + | 0 & 0 & 1 | ||
| + | \end{bmatrix} | ||
| + | = | ||
| + | \begin{bmatrix} | ||
| + | 1 & 0 & 0 \\ | ||
| + | 3 & 1 & 0 \\ | ||
| + | 2 & \frac{1}{2} & 1 | ||
| + | \end{bmatrix} | ||
| + | \times | ||
| + | \begin{bmatrix} | ||
| + | 1 & 3 & -2 \\ | ||
| + | 0 & -4 & 12 \\ | ||
| + | 0 & 0 & 1 | ||
| + | \end{bmatrix} | ||
| + | </math> | ||
| + | |||
| + | Given L and U we can solve '''Ax'''='''b''' by first solving '''Ly'''='''b''', where: | ||
| + | |||
| + | :<math> | ||
| + | \begin{bmatrix} | ||
| + | 1 & 0 & 0 \\ | ||
| + | 3 & 1 & 0 \\ | ||
| + | 2 & \frac{1}{2} & 1 | ||
| + | \end{bmatrix} | ||
| + | \begin{bmatrix}y_1\\y_2\\y_3\end{bmatrix} | ||
| + | = | ||
| + | \begin{bmatrix}5\\7\\8\end{bmatrix} | ||
| + | </math> | ||
| + | |||
| + | As L is unit lower triangular, we can do this by easily by forward substitution: | ||
| + | |||
| + | :<math> | ||
| + | \begin{array}{rcl} | ||
| + | y_1 & = & 5 \\ | ||
| + | 3 \times y_1 + y_2 & = & 7 \\ | ||
| + | y_2 & = & 7 - 15 \\ | ||
| + | y_2 & = & -8 \\ | ||
| + | 10 - 4 + y_3 & = & 8 \\ | ||
| + | y_3 & = & 12 - 10 \\ | ||
| + | y_3 & = & 2 | ||
| + | \end{array} | ||
| + | </math> | ||
| + | |||
| + | Once we know y, we can solve '''Ux'''='''y''': | ||
| + | |||
| + | :<math> | ||
| + | \begin{bmatrix} | ||
| + | 1 & 3 & -2 \\ | ||
| + | 0 & -4 & 12 \\ | ||
| + | 0 & 0 & 1 | ||
| + | \end{bmatrix} | ||
| + | \begin{bmatrix}x_1\\x_2\\x_3\end{bmatrix} | ||
| + | = | ||
| + | \begin{bmatrix}5\\-8\\2\end{bmatrix} | ||
| + | </math> | ||
| + | |||
| + | This time easily by backward substitution: | ||
| + | |||
| + | :<math> | ||
| + | \begin{array}{rcl} | ||
| + | x_3 & = & 2 \\ | ||
| + | 24 - 4 \times x_2 & = & -8 \\ | ||
| + | 32 & = & 4x_2 \\ | ||
| + | x_2 & = & 8 \\ | ||
| + | -4 + 24 + x_1 & = & 5 \\ | ||
| + | x_1 & = & 9 - 24 \\ | ||
| + | x_1 & = & -15 | ||
| + | \end{array} | ||
| + | </math> | ||
| + | |||
| + | Computing the LU decomposition of an <math>n \times n</math> system of equations has a computational cost of <math>O(n^3)</math> operations, whereas the forward and backward substitution steps have a cost of <math>O(n^2)</math> operations. '''Thus the cost of solution is dominated by the LU decomposition'''. | ||
| + | |||
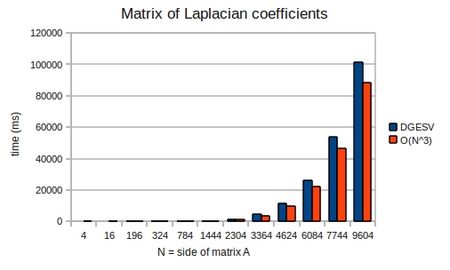
| + | To demonstrate that the solution does indeed take <math>O(n^3)</math> operations, I ran '''mxm-laplacian.exe''' on my desktop PC for varying sizes of N and could plot the following from the results: | ||
| + | |||
| + | [[Image:dgesv-laplacian-timings.jpg|450px|centre|thumbnail|Time taken by LAPACK's degsv routine for a matrix of laplacian coefficents.]] | ||
| + | |||
| + | If we look once again at '''dgesv-example.f90''' and set: | ||
| + | |||
| + | <pre>logical, parameter :: verbose=.true.</pre> | ||
| + | |||
| + | (and recompile and run) we can see the contents of A after the call to dgesv(). It now contains an encoding of the LU decomposition (the unit leading diagonal from L is assumed): | ||
| + | |||
| + | <pre> | ||
| + | LU decomposition as encoded in matrix A: | ||
| + | 3.00000000000000 5.00000000000000 6.00000000000000 | ||
| + | 0.333333333333333 1.33333333333333 -4.00000000000000 | ||
| + | 0.666666666666667 0.500000000000000 1.00000000000000 | ||
| + | |||
| + | ..and the IPIV vector: | ||
| + | 2 2 3 | ||
| + | </pre> | ||
| + | |||
| + | This is all well and good, but '''hang-on a cotton pickin' minute'''!, the matrix U is not as we expected: | ||
| + | |||
| + | :<math> | ||
| + | \begin{bmatrix} | ||
| + | 3 & 5 & 6 \\ | ||
| + | 0 & \frac{4}{3} & -4 \\ | ||
| + | 0 & 0 & 1 | ||
| + | \end{bmatrix} | ||
| + | \neq | ||
| + | \begin{bmatrix} | ||
| + | 1 & 3 & -2 \\ | ||
| + | 0 & -4 & 12 \\ | ||
| + | 0 & 0 & 1 | ||
| + | \end{bmatrix} | ||
| + | </math> | ||
| + | |||
| + | Why not? The answer lies the in contents of the IPIV. The elements along the leading diagonal are known as '''pivots'''. If any pivots are small, our algorithm will become unstable. We may exchange rows (known as '''partial pivoting''') in A without changing the solution. We can read the elements of IPIV as a set of instructions for manipulating each row of the original matrix. | ||
| + | |||
| + | For example, if the first element of IPIV is a 1, then we read this as: | ||
| + | * exchange row 1 with row 1 (i.e. no change--a null operation) | ||
| + | but if the first element of IPIV were a 2, then this means: | ||
| + | * exchange row 1 with row 2 (i.e. swap round the firs two rows) | ||
| + | |||
| + | In the case of our example program, the following sequence of actions were performed: | ||
| + | # row 1 was exchanged with row 2 | ||
| + | # row 2 was exchanged with row 2 | ||
| + | # row 3 was exchanged with row 3 | ||
| + | |||
| + | We can satisfy ourselves that all is well, by factorising this permuted matrix: | ||
| + | |||
| + | :<math> | ||
| + | \begin{bmatrix} | ||
| + | 1 & 0 & 0 \\ | ||
| + | 0 & 1 & 0 \\ | ||
| + | 0 & \frac{1}{2} & 1 | ||
| + | \end{bmatrix} | ||
| + | \times | ||
| + | \begin{bmatrix} | ||
| + | 1 & 0 & 0 \\ | ||
| + | \frac{1}{3} & 1 & 0 \\ | ||
| + | \frac{2}{3} & 0 & 1 | ||
| + | \end{bmatrix} | ||
| + | \times | ||
| + | \begin{bmatrix} | ||
| + | 3 & 5 & 6 \\ | ||
| + | 0 & \frac{4}{3} & -4 \\ | ||
| + | 0 & 0 & 1 | ||
| + | \end{bmatrix} | ||
| + | = | ||
| + | \begin{bmatrix} | ||
| + | 3 & 5 & 6 \\ | ||
| + | 1 & 3 & -2 \\ | ||
| + | 2 & 4 & 3 | ||
| + | \end{bmatrix} | ||
| + | </math> | ||
| + | |||
| + | ==Theory: Parabolic Equations & Forward (Explicit) Euler== | ||
| + | |||
| + | Let's look at how to solve a different class of equation, again using finite difference techniques. | ||
| + | |||
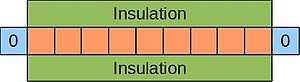
| + | An example of a parabolic equation is the 1D heat equation, which describes the evolution of temperature at various points in a thin, insulated bar over time. Imagine such a bar where the ends are kept at 0°C: | ||
| + | |||
| + | [[Image:Insulated-rod.jpg|300px|centre|thumbnail|A thin, insulated rod. Ends kept at 0°C.]] | ||
| + | |||
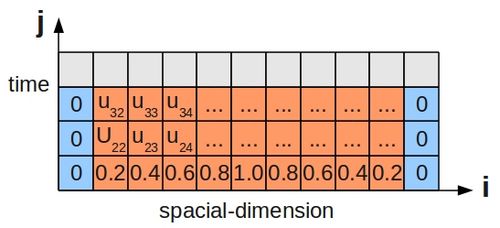
| + | Since this scenario involves the passage of time, just stating the '''boundary conditions''' is not enough, and we must also specify some '''initial condition''' for the temperature along the bar's length. This is shown below for a bar divided up into grid cells, as before: | ||
| + | |||
| + | [[Image:1DHeatEquFiniteDiff.jpg|500px|centre|thumbnail|Finite Difference Grid for 1D Heat Equation]] | ||
| + | |||
| + | The 1D heat equation is: | ||
| + | |||
| + | :<math>\frac{\partial u}{\partial t}=\alpha \frac{\partial^2u}{\partial x^2} </math> | ||
| + | |||
| + | where <math>\alpha</math>--the thermal diffusivity--is set to 1 for simplicity. ''One way'' to rewrite this as a difference equation (using a forward difference for the LHS and a central difference for the RHS*) is: | ||
| + | |||
| + | :<math>\frac{u^{j+1}_i - u^j_i }{k} = \frac{u^j_{i+1} - 2u^j_i + u^j_{i-1}}{h^2}</math> | ||
| + | |||
| + | and rearrange to solve for <math>u^{j+1}_i</math>: | ||
| + | |||
| + | :<math>u^{j+1}_i = u^j_i + r(u^j_{i+1} - 2u^j_i + u^j_{i-1})</math> | ||
| + | |||
| + | :<math>u^{j+1}_i = ru^j_{i+1} + (1- 2r)u^j_i + ru^j_{i-1}</math> | ||
| + | |||
| + | where: | ||
| + | |||
| + | <math>r=\frac{k}{h^2}</math> | ||
| + | |||
| + | This time, we see that the appropriate stencil is: | ||
| + | |||
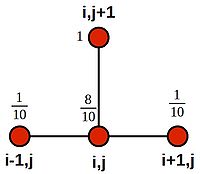
| + | [[Image:forward-euler-stencil.jpg|200px|thumbnail|centre|Stencil for the (explicit) forward Euler method]] | ||
| + | |||
| + | If we set <math>h=\frac{1}{10}</math> and <math>k=\frac{1}{1000}</math>, then <math>r=\frac{1}{10}</math>, and the equation becomes: | ||
| + | |||
| + | :<math>u^{j+1}_i = u^j_i + \frac{1}{10}(u^j_{i+1} - 2u^j_i + u^j_{i-1})</math> | ||
| + | |||
| + | :<math>u^{j+1}_i = \frac{1}{10}(u^j_{i+1} + 10u^j_i - 2u^j_i + u^j_{i-1})</math> | ||
| + | |||
| + | :<math>u^{j+1}_i = \frac{1}{10}(u^j_{i+1} + 8u^j_i + u^j_{i-1})</math> | ||
| + | |||
| + | Which we can illustrate using weightings on the stencil nodes: | ||
| + | |||
| + | [[Image:Forward-euler-stencil-weights.jpg|200px|thumbnail|centre|Stencil for the (explicit) forward Euler method showing node weights]] | ||
| + | |||
| + | Forward Euler is as an '''explicit''' method, as one unknown value (<math>u^{j+1}_i</math>) is expressed directly in terms of known values (all values of u at time j are known). | ||
| + | |||
| + | We could implement a simple marching algorithm, where we centre the stencil over a set of known values at time j, and may calculate the unknown temperature for grid cell i at time j+1. However, it would be better to group together all the calculations for a particular time step into a system of simultaneous equations (for our example 1d heat problem): | ||
| + | |||
| + | :<math>u_{22} = \frac{1}{10}(0) + \frac{8}{10}(0.2) + \frac{1}{10}(0.4)</math> | ||
| + | |||
| + | :<math>u_{23} = \frac{1}{10}(0.2) + \frac{8}{10}(0.4) + \frac{1}{10}(0.6)</math> | ||
| + | |||
| + | :<math>u_{24} = \frac{1}{10}(0.4) + \frac{8}{10}(0.6) + \frac{1}{10}(0.8)</math> | ||
| + | |||
| + | :<math>\cdots</math> | ||
| + | |||
| + | and then re-write in matrix form: | ||
| + | |||
| + | :<math> | ||
| + | \begin{bmatrix}u_{22} \\ u_{23} \\ u_{24} \\\vdots \end{bmatrix} | ||
| + | = | ||
| + | \begin{bmatrix} | ||
| + | 0.1 & 0.8 & 0.1 & 0 & 0 & \cdots & 0 \\ | ||
| + | 0 & 0.1 & 0.8 & 0.1 & 0 & \cdots & 0 \\ | ||
| + | \vdots & & & & & & \vdots \\ | ||
| + | & & \cdots & 0 & 0.1 & 0.8 & 0.1 \\ | ||
| + | \end{bmatrix} | ||
| + | \times | ||
| + | \begin{bmatrix}0 \\ 0.2 \\ 0.4 \\ \vdots \\ 0.2 \\ 0 \end{bmatrix} | ||
| + | </math> | ||
| + | |||
| + | as the computational efficiency of a matrix vector multiplication will be hard to better. Note that we can write the above piece of linear algebra in a general form: | ||
| + | |||
| + | :<math> | ||
| + | \begin{bmatrix}u_{22} \\ u_{23} \\ u_{24} \\ \vdots \end{bmatrix} | ||
| + | = | ||
| + | \begin{bmatrix} | ||
| + | r & 1-2r & r & 0 & 0 & \cdots & 0 \\ | ||
| + | 0 & r & 1-2r & r & 0 & \cdots & 0 \\ | ||
| + | \vdots & & & & & & \vdots \\ | ||
| + | & & \cdots & 0 & r & 1-2r & r \\ | ||
| + | \end{bmatrix} | ||
| + | \times | ||
| + | \begin{bmatrix}u_{11} \\ u_{12} \\ u_{13} \\ u_{14} \\ \vdots \end{bmatrix} | ||
| + | </math> | ||
| + | |||
| + | (*The two difference schemes for either side of the equation were chosen so that the approximation error characteristics for either side are approximately balanced and so the errors on one side or the other do not begin to dominate the calculations.) | ||
| + | |||
| + | ==Theory: Solution Stability and the CFL Condition== | ||
| + | |||
| + | The Forward Euler scheme has the benefit of simplicity, but is unstable for large time steps. The CFL (Courant–Friedrichs–Lewy) condition states that the following must hold: | ||
| + | |||
| + | :<math>r=\frac{k}{h^2} \le 0.5 </math> | ||
| + | |||
| + | for stability. | ||
| + | |||
| + | ==Theory: Parabolic Equations & Backward (Implicit) Euler== | ||
| + | |||
| + | Another way to formulate our finite difference equations for the very same problem is to use backward-differences instead of forward-differences. The backward Euler scheme requires more computation per time-step than the forward scheme, but has the distinct advantage that it is '''unconditionally stable''' with regard to the relative size between the time and spatial step sizes; i.e. we are not limited to <math>r \le 0.5</math> when we use the backward Euler scheme. | ||
| + | |||
| + | We will use a central-difference on the RHS, as before, but shifting the time index to <math>j+1</math>. This will allow us to formulate a backward-difference on the RHS between the time indices <math>j</math> and <math>j+1</math>: | ||
| + | |||
| + | :<math>\frac{u^{j+1}_i - u^j_i }{k} = \frac{u^{j+1}_{i+1} - 2u^{j+1}_i + u^{j+1}_{i-1}}{h^2}</math> | ||
| + | |||
| + | :<math>u^{j+1}_i - u^j_i = \frac{k}{h^2}(u^{j+1}_{i+1} - 2u^{j+1}_i + u^{j+1}_{i-1})</math> | ||
| + | |||
| + | :<math>u^{j+1}_i - u^j_i = r(u^{j+1}_{i+1} - 2u^{j+1}_i + u^{j+1}_{i-1})</math> | ||
| + | |||
| + | :<math>u^{j+1}_i - u^j_i = ru^{j+1}_{i+1} - 2ru^{j+1}_i + ru^{j+1}_{i-1}</math> | ||
| + | |||
| + | :<math>u^{j+1}_i - ru^{j+1}_{i+1} + 2ru^{j+1}_i - ru^{j+1}_{i-1} = u^j_i</math> | ||
| + | |||
| + | :<math>u^j_i = - ru^{j+1}_{i+1} + (1 + 2r)u^{j+1}_i - ru^{j+1}_{i-1}</math> | ||
| + | |||
| + | If we again choose <math>h=\frac{1}{10}</math> and <math>k=\frac{1}{1000}</math>, such that <math>r=\frac{k}{h^2}=\frac{1}{10}</math>, as before, we end up with the following system of equations when we consider all values of <math>i</math> for our first time-step--from <math>j</math> to <math>j+1</math>--where we know values of all the cells at time <math>j</math>: | ||
| + | |||
| + | :<math>0.2 = -\frac{1}{10}(0) + \frac{12}{10}(u_{22}) -\frac{1}{10}(u_{23})</math> | ||
| + | |||
| + | :<math>0.4 = -\frac{1}{10}(u_{22}) + \frac{12}{10}(u_{23}) -\frac{1}{10}(u_{24})</math> | ||
| + | |||
| + | :<math>0.6 = -\frac{1}{10}(u_{23}) + \frac{12}{10}(u_{24}) -\frac{1}{10}(u_{25})</math> | ||
| + | :<math>\cdots</math> | ||
| + | |||
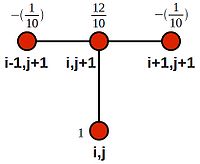
| + | We can see that this is akin to looping over the domain with the following stencil: | ||
| + | |||
| + | [[Image:Backward-euler-stencil-weighted.jpg|200px|thumbnail|centre|Stencil for the (implicit) backward Euler method showing node weights]] | ||
| + | |||
| + | Rearranging, so that we have only constants on the RHS: | ||
| + | |||
| + | :<math>\frac{1}{10}(0) + \frac{12}{10}(u_{22}) -\frac{1}{10}(u_{23}) = 0.2 + \frac{1}{10}(0)</math> | ||
| + | |||
| + | :<math>\frac{1}{10}(u_{22}) + \frac{12}{10}(u_{23}) -\frac{1}{10}(u_{24}) = 0.4</math> | ||
| + | |||
| + | :<math>\frac{1}{10}(u_{23}) + \frac{12}{10}(u_{24}) -\frac{1}{10}(u_{25}) = 0.6</math> | ||
| + | :<math>\cdots</math> | ||
| + | |||
| + | We can re-write this system of equations in matrix form: | ||
| + | |||
| + | :<math> | ||
| + | \begin{bmatrix} | ||
| + | 1.2 & -0.1 & 0 & 0 & \cdots & 0 \\ | ||
| + | -0.1 & 1.2 & -0.1 & 0 & \cdots & 0 \\ | ||
| + | \vdots & & & & & \vdots \\ | ||
| + | & & \cdots & 0 & -0.1 & 1.2 \\ | ||
| + | \end{bmatrix} | ||
| + | \times | ||
| + | \begin{bmatrix} u_{22} \\ u_{23} \\ u_{24} \\\vdots \end{bmatrix} | ||
| + | = | ||
| + | \begin{bmatrix} 0.2 + 0.1(0) \\ 0.4 \\ \vdots \\ 0.2 +0.1(0) \end{bmatrix} | ||
| + | </math> | ||
| + | |||
| + | The general form is: | ||
| + | |||
| + | :<math> | ||
| + | \begin{bmatrix} | ||
| + | 1+2r & -r & 0 & 0 & \cdots & 0 \\ | ||
| + | -r & 1+2r & -r & 0 & \cdots & 0 \\ | ||
| + | \vdots & & & & & \vdots \\ | ||
| + | & & \cdots & 0 & -r & 1+2r \\ | ||
| + | \end{bmatrix} | ||
| + | \times | ||
| + | \begin{bmatrix} u_{22} \\ u_{23} \\ u_{24} \\\vdots \end{bmatrix} | ||
| + | = | ||
| + | \begin{bmatrix} u_{12} + r(u_{11}) \\ u_{13} \\ \vdots \\ u_{1n-1} + r(u_{1n}) \end{bmatrix} | ||
| + | </math> | ||
| + | |||
| + | ==Theory: Parabolic Equations & Crank-Nicolson== | ||
| + | |||
| + | Now, forward and backward Euler schemes are by far from the only ways in which to approach this 1D heat problem. Yet another scheme is the to create finite difference equations using the Crank-Nicolson stencil: | ||
| + | |||
| + | [[Image:crank-nicolson-stencil.jpg|200px|thumbnail|centre|Stencil for the (implicit) Crank-Nicolson method]] | ||
| + | |||
| + | The stencil amounts to an average of both the forward and backward Euler methods. The resulting difference equation is: | ||
| + | |||
| + | :<math>\frac{u^{j+1}_i - u^j_i}{k} = \frac{1}{2}(\frac{u^{j+1}_{i+1} - 2u^{j+1}_{i} + u^{j+1}_{i-1}}{h^2} + \frac{u^j_{i+1} - 2u^j_i + u^j_{i-1}}{h^2}).</math> | ||
| + | |||
| + | Which we may rearrange to become: | ||
| + | |||
| + | :<math>u^{j+1}_i - u^j_i = \frac{1}{2}\frac{k}{h^2}(u^{j+1}_{i+1} - 2u^{j+1}_i + u^{j+1}_{i-1} + u^j_{i+1} - 2u^j_i + u^j_{i-1})</math> | ||
| + | |||
| + | :<math>2(u^{j+1}_i - u^j_i) = r(u^{j+1}_{i+1} - 2u^{j+1}_i + u^{j+1}_{i-1} + u^j_{i+1} - 2u^j_i + u^j_{i-1})</math>, | ||
| + | |||
| + | where <math>r=\frac{k}{h^2}</math>. | ||
| + | |||
| + | Collecting the unknowns (values of u at the next timestep, <math>j+1</math>) to the LHS and the knowns (values of u at current timestep, <math>j</math>), we get: | ||
| + | |||
| + | :<math>2u^{j+1}_i + 2ru^{j+1}_i - ru^{j+1}_{i+1} - ru^{j+1}_{i-1} = 2u^j_i +ru^j_{i+1} -2ru^j_i + ru^j_{i-1}</math> | ||
| + | |||
| + | :<math>(2+2r)u^{j+1}_i - ru^{j+1}_{i+1} - ru^{j+1}_{i-1} = (2-2r)u^j_i +ru^j_{i+1} + ru^j_{i-1}</math>. | ||
| + | |||
| + | If we choose <math>h=1/10</math> and <math>k=1/100</math> (note that the time-step is 10x larger than than chosen for the forward Euler example), then <math>r=1</math>, and we get: | ||
| + | |||
| + | :<math>-u_{i-1}^{j+1} +4u_i^{j+1} -u_{i+1}^{j+1} = u_{i-1}^j + u_{i+1}^j</math>. | ||
| + | |||
| + | We can again create a system of linear equations for all the cells j along the bar at a given time step i: | ||
| + | |||
| + | :<math>-0 + 4u_{22} - u_{23} = 0 + 0.4</math> | ||
| + | |||
| + | :<math>-u_{22} + 4u_{23} - u_{24} = 0.2 + 0.6</math> | ||
| + | |||
| + | :<math>-u_{23} + 4u_{24} - u_{25} = 0.4 + 0.8</math> | ||
| + | |||
| + | :<math>\cdots</math> | ||
| + | |||
| + | We have <math>j</math> unknowns and <math>j</math> equations. Then we can again translate into matrix notation and solve using a library linear algebra routines: | ||
| + | |||
| + | :<math> | ||
| + | \begin{bmatrix} | ||
| + | -4 & 1 & 0 & 1 & 0 & 0 & \cdots & 0 \\ | ||
| + | 1 & -4 & 1 & 0 & 1 & 0 & \cdots & 0 \\ | ||
| + | \vdots & & & & & & & \vdots \\ | ||
| + | & \cdots & & 0 & 1 & 0 & 1 & -4\\ | ||
| + | \end{bmatrix} | ||
| + | \times | ||
| + | \begin{bmatrix} u_{22} \\ u_{23} \\ u_{24} \\\vdots \end{bmatrix} | ||
| + | = | ||
| + | \begin{bmatrix} 0 + 0.4 \\ 0.2 + 0.6 \\ \vdots \\ 0 + 0.4 \end{bmatrix} | ||
| + | </math> | ||
| + | |||
| + | In general: | ||
| + | |||
| + | :<math> | ||
| + | \begin{bmatrix} | ||
| + | -(2+2r) & r & 0 & r & 0 & 0 & \cdots & 0 \\ | ||
| + | r & -(2+2r) & r & 0 & r & 0 & \cdots & 0 \\ | ||
| + | \vdots & & & & & & & \vdots \\ | ||
| + | & \cdots & & 0 & r & 0 & r & -(2+2r)\\ | ||
| + | \end{bmatrix} | ||
| + | \times | ||
| + | \begin{bmatrix} u_{22} \\ u_{23} \\ u_{24} \\\vdots \end{bmatrix} | ||
| + | = | ||
| + | \begin{bmatrix} ru_{11} + (2-2r)u_{12} + ru_{13} \\ \vdots \end{bmatrix} | ||
| + | </math> | ||
| + | |||
| + | ==Practice: Solving Parabolic Problems using LAPACK== | ||
| + | |||
| + | In our second example, we'll see 3 different schemes for solving a parabolic problem--forward Euler, backward Euler and Crank-Nicolson. The forward Euler scheme is explicit and so no system of linear equations is solved. It is a appealingly simple scheme, but can suffer from unstable solutions under certain conditions. In both the backward Euler and Crank-Nicolson schemes, we will see the (repeated) use of a pre-computed LU decomposition when solving a parabolic equation. | ||
| + | |||
| + | <pre> | ||
| + | cd ../example2 | ||
| + | </pre> | ||
| + | |||
| + | In '''simple-crank-nicolson.f90''' we see that the initial call to '''dgesv''' replaces the entries in the matrix A with an encoding of the LU decompostion (the unit diagonal is assumed). For subsequent timesteps we can thus call '''dgetrs''' instead: | ||
| + | |||
| + | <source lang="fortran"> | ||
| + | do ii=2,4 | ||
| + | ... | ||
| + | call dgetrs('N', N, NRHS, A, LDA, IPIV, B, LDB, INFO) | ||
| + | ... | ||
| + | end do | ||
| + | </source> | ||
| + | |||
| + | where we have remembered to take account of the boundary conditions at each time-step too: | ||
| + | |||
| + | <source lang="fortran"> | ||
| + | do ii=2,4 | ||
| + | ... | ||
| + | ! copy B into B1, so that we can recompute for | ||
| + | ! the boundary conditions again. | ||
| + | B1 = B | ||
| + | |||
| + | ! Need to recalculate entries for the vector of known | ||
| + | ! values--held in B--given the solution at the current | ||
| + | ! timestep--held in B1. | ||
| + | ! *NB* the loops below are only valid for the case r=1. | ||
| + | ! LHS | ||
| + | B(1,1) = 0 + B1(2,1) | ||
| + | ! RHS | ||
| + | B(N,1) = 0 + B1(N-1,1) | ||
| + | ! middle bit | ||
| + | do jj=2,N-1 | ||
| + | B(jj,1) = B1(jj-1,1) + B1(jj+1,1) | ||
| + | end do | ||
| + | ... | ||
| + | end do | ||
| + | </source> | ||
| + | |||
| + | ---- | ||
| + | |||
| + | '''Exercises''' | ||
| + | |||
| + | * Experiment with values of <math>r=\frac{k}{h^2}</math> for the 3 schemes, so as to highlight the effects of the CFL condition. | ||
| + | |||
| + | ==Practice: Banded Matrices== | ||
| + | |||
| + | In all my examples, thus far, I've called routines from the '''dge'''xxx series offered by LAPACK. The '''d''' stands for '''d'''ouble and the '''ge''' for '''ge'''nerel purpose. For banded matrices, LAPACK offers a much more efficient format and routines to match. Take a look at http://www.netlib.org/lapack/double/ for a list of driver routines using matrices of double precision numbers, or http://www.netlib.org/lapack for more documentation in general. | ||
| + | |||
| + | ==Practice: PETSc & Iterative solutions and sparse matrices== | ||
| + | |||
| + | In order to compute the solution of a linear system via the direct method of an LU-decomposition, we know that it will cost <math>O(n^3)</math> operations. Can we compute a solution more cheaply? The class of iterative methods strive to compute a cheaper solution by first guessing a value for the solution vector <math>x</math> and then iteratively refining it. Thus we will get a series of (hopefully!) improving approximations: | ||
| + | |||
| + | :<math> | ||
| + | x^0, x^1, x^2, x^3, \cdots, x^j \rightarrow x | ||
| + | </math> | ||
| + | |||
| + | By way of an illustration of such a method, consider splitting matrix A into 3 others: | ||
| + | |||
| + | :<math> | ||
| + | A = L + D + U | ||
| + | </math> | ||
| + | |||
| + | Note the summation, and so the L and the U matrices are '''not the same as those considered in the LU-decomposition''' used by the direct approach. With this split, '''Ax'''='''b''', becomes: | ||
| + | |||
| + | :<math> | ||
| + | Lx + Dx + Ux = b | ||
| + | </math> | ||
| + | |||
| + | The game now is to replace some occurrences of <math>x</math> by <math>x^k</math> and others by <math>x^{k+1}</math>, and thereby obtaining a relationship which tells us how to get an approximation at one iteration step, from the approximation at the previous step. If the resulting iterative scheme converges, then we're in business! | ||
| + | |||
| + | One classic form of such an iteration is the Jacobi method (which is useful when A is diagonally dominant): | ||
| + | |||
| + | :<math> | ||
| + | Lx^j + Dx^{j+1} + Ux^j = b | ||
| + | </math> | ||
| + | |||
| + | :<math> | ||
| + | x^{j+1} = D^{-1}(b - (L + U)x^j) | ||
| + | </math> | ||
| + | |||
| + | To take a look at a illustrative implementation of the Jacobi method using LAPACK: | ||
| + | |||
| + | <pre> | ||
| + | cd ../example3 | ||
| + | make | ||
| + | </pre> | ||
| + | |||
| + | and take a look at '''jacobi-example.f90'''. To run the executable, type: | ||
| + | |||
| + | <pre> | ||
| + | ./jacobi-example.exe | ||
| + | </pre> | ||
| + | |||
| + | Happily, however, we do not need to write our own iterative schemes, as the good folk at Argonne have written the '''PETSc''' library. | ||
| + | |||
| + | http://www.mcs.anl.gov/petsc/petsc-as/ | ||
| + | |||
| + | Take a look inside '''ex1f.F''' to see how to solve our laplacian example using PETSc. To run the program you will obviously need PETSc installed on the computer you are using. If you have, then edit Makefile and set PETSC_DIR to the appropriate value, and then type: | ||
| + | |||
| + | <pre> | ||
| + | make ex1f | ||
| + | qsub mpi_submit | ||
| + | </pre> | ||
| + | |||
| + | Look in the file called '''OUT''' for the results of running PETSc. | ||
| + | |||
| + | We can again plot a graph of timings given varying N: | ||
| + | |||
| + | [[Image:gmres-vs-LU.jpg|450px|centre|thumbnail|Comparing the time taken by an iterative method to that taken by LAPACK's LU decomposition for a sparse matrix.]] | ||
| + | |||
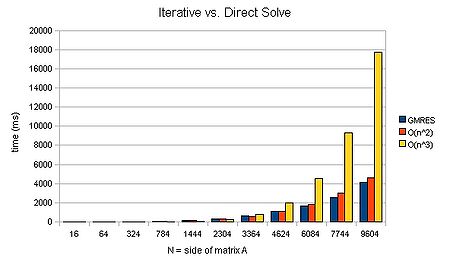
| + | We can see that an iterative method can scale approximately <math>O(n^2)</math>. For large matrices, this can mean that a solution is found several orders of magnitude faster. Now that is a speed-up worth having! It's liking buying a supercomputer than runs several orders of magnitude faster than your old one.. | ||
| + | |||
| + | Better yet, you can try different schemes for iterative solution, just by varying some command-line arguments. For example: | ||
| + | |||
| + | <pre> | ||
| + | ./ex1f -ksp_type bcgs -pc_type jacobi | ||
| + | </pre> | ||
| + | |||
| + | will invoke the biconjugate gradient stabalised scheme together with a jacobi preconditioner matrix. (This in fact runs faster than the GMRES method for the above case--808ms for N=9694. Note, however, that it cannot scale better than <math>O(N^2)</math>.) For those interested, the PETSc manual is the best next port of call for more details on the various methods made available by the package. | ||
| + | |||
| + | For further examples see INSTALL_DIR/petsc-3.1-p1/src/ksp/ksp/examples/tutorials. | ||
| + | |||
| + | =Other Numerical Methods= | ||
| + | |||
| + | We have looked at the ''finite difference'' method (FDM) above, but other (more mathsy) approaches exist, such as the ''finite element'' and ''finite volume'' methods. Each approach has its pros and cons. For example, the FDM is limited to regular grids spread over rectangular domains, but is easy to program. In contrast the FEM is applicable to intricately shaped domains and the resolution of the individual elements can be varied, but is requires more involved programming and a deeper level of understand of the relevant mathematics. It should also be noted, however, that under certain circumstances, the FEM and FVM boil down to be ''exactly the same'' as the FDM, even though we've approached the task of solving a PDE from a different direction. | ||
| + | |||
| + | One thing which is ''always'' common to the approaches is that ''the problem boils down to a system of linear equations to solve, and that a library of linear algebra routines will be the workhorse when finding solutions to PDEs on a computer''. Repeat after me, "a fast linear algebra library is key!" | ||
| + | |||
| + | ==Theory: The Finite Element Method (FEM)== | ||
| + | |||
| + | The general approach here is to create a potential function from our PDE and to find a solution to it via minimising the potential function. Key steps include: | ||
| + | |||
| + | * Discritisation of the original PDE, often by proposing that we can approximate the solution using a linear sum of (polynomial) basis functions. | ||
| + | * Create the potential function by invoking the method of weighted residuals, and in particular the Galerkin specialisation of this approach. | ||
| + | * Use whatever algebraic manipulation and integration (possibly numerical) is required to create a system of linear equations. | ||
| + | * Solve said system of linear equations and in doing so, solve the original PDE. | ||
| + | |||
| + | For example, consider again Poisson's equation over a 2-dimensional region, <math>R</math>: | ||
| + | |||
| + | :<math>\nabla^2u=f(x,y)</math> | ||
| + | |||
| + | where <math>u=0</math> at the boundary of the domain. | ||
| + | |||
| + | Next, applying the method of weighted residuals, we multiply both sides by weighting functions <math>\phi_i</math>, and take integrals over the domain: | ||
| + | |||
| + | :<math> \iint\limits_R \nabla^2u\phi_i\, dA = \iint\limits_R f\phi_i\, dA </math> | ||
| + | |||
| + | We can apply integration-by-parts to the LHS (<math>\phi_i\nabla^2u</math>), for example: | ||
| + | |||
| + | :<math>\int \phi_i\nabla^2 u = \phi_i\nabla u - \int \nabla\phi_i \cdot \nabla u</math> | ||
| + | |||
| + | Differentiating both sides: | ||
| + | |||
| + | :<math>\phi_i\nabla^2 u = \nabla \cdot (\phi_i\nabla u) - \nabla\phi_i \cdot \nabla u</math> | ||
| + | |||
| + | So inserting this into our equation for weighted residuals, above: | ||
| + | |||
| + | :<math> \iint\limits_R [\nabla \cdot (\phi_i\nabla u) - \nabla\phi_i \cdot \nabla u]\, dA = \iint\limits_R f\phi_i\, dA </math> | ||
| + | |||
| + | and rearranging, we get: | ||
| + | |||
| + | :<math> \iint\limits_R \nabla\phi_i \cdot \nabla u\, dA = \iint\limits_R \nabla \cdot (\phi_i\nabla u)\, dA - \iint\limits_R f\phi_i\, dA </math> | ||
| + | |||
| + | We can see that the 1st term on the RHS is a divergence(the divergence of a vector field <math>F</math>, <math>div F = \nabla \cdot F</math>), and so we can apply Gauss' Divergence Theorem which states, for the region <math>R</math> bounded by the curve s: | ||
| + | |||
| + | :<math>\iint\limits_R \nabla \cdot F\, dA = \oint\limits_s F \cdot n\, ds </math> | ||
| + | |||
| + | where <math>n</math> is the unit vector normal to the surface. | ||
| + | |||
| + | and so, | ||
| + | |||
| + | :<math> \iint\limits_R \nabla\phi_i \cdot \nabla u\, dA = \oint\limits_s \phi_i\nabla u \cdot n\, ds - \iint\limits_R f\phi_i\, dA </math> | ||
| + | |||
| + | but our homogeneous boundary conditions also tell us that <math>u</math> is equal to zero at the boundary and so the 1st term on the RHS goes to zero: | ||
| + | |||
| + | :<math> \iint\limits_R \nabla\phi_i \cdot \nabla u\, dA = - \iint\limits_R f\phi_i\, dA </math> | ||
| + | |||
| + | So far, all of the operations above have been done on our unaltered, continuous differential equation. At this point, let's introduce the discritisation--and hence the approximation--that we need to make to work on a digital computer. We intend to approximate the solution <math>u(x,y)</math> using a linear combination of basis functions: | ||
| + | |||
| + | :<math>U(x,y) = \sum_{j=1}^m U_j\phi_j(x,y)</math> | ||
| + | |||
| + | The Galerkin method is being used here, as our basis functions are the same as the weighting functions used above. | ||
| + | |||
| + | :<math> \sum_{j=1}^m U_j \iint\limits_R \nabla\phi_i \cdot \nabla\phi_j \, dA = - \iint\limits_R f\phi_i\, dA</math> | ||
| + | |||
| + | At this point, we can introduce a symmetric 'stiffness matrix' with entries: | ||
| + | |||
| + | :<math>K_{ij} = \iint\limits_R \nabla\phi_i \cdot \nabla\phi_j \, dA</math> | ||
| + | |||
| + | and a vector '''F''', with entries: | ||
| + | |||
| + | :<math>F_i = - \iint\limits_R f\phi_i\, dA</math> | ||
| + | |||
| + | and thus our <math>m</math> equations: | ||
| + | |||
| + | :<math>\sum_{j=1}^m K_{ij}U_j = F_i </math> | ||
| + | |||
| + | which, we see, yet again is a linear system of equations of the form '''Ax'''='''b'''. Cue a call to a linear algebra library! | ||
| + | |||
| + | If we use linear basis functions over our domain, we will end up with the same system of linear equations to solve as we would get following the FDM: | ||
| + | |||
| + | [[Image:FE-linear-basis.jpg|300px|thumbnail|centre|Linear basis functions interpolating over triangular elements.]] | ||
| + | |||
| + | Rather than trying to write an example program to demonstrate the FEM, it makes much more sense to look at a freely available FEM package, such as Elmer (http://www.csc.fi/english/pages/elmer). Below are images obtained by running one of the examples included with Elmer: | ||
| + | |||
| + | [[Image:Elmer.jpg|500px|centre|thumbnail|A FEM mesh used by Elmer to solve a steady state heat problem.]] | ||
| + | [[Image:Elmer-solution.jpg|500px|centre|thumbnail|The solution to the steady state heat problem as computed by Elmer.]] | ||
| + | |||
| + | =Other Linear Algebra Packages= | ||
| + | |||
| + | ==Practice: PLASMA for Multicore Architectures== | ||
| + | |||
| + | http://icl.cs.utk.edu/plasma/index.html | ||
| + | |||
| + | <pre> | ||
| + | cd ../example4 | ||
| + | make | ||
| + | ./mxm-laplace-plasma.exe | ||
| + | </pre> | ||
| + | |||
| + | For further examples, see e.g. INSTALL_DIR/plasma-installer_2.3.1/build/plasma_2.3.1. | ||
| + | |||
| + | ==Practice: MAGMA to use GPUs in Heterogeneous Systems== | ||
| + | |||
| + | http://icl.cs.utk.edu/magma/index.html | ||
| + | |||
| + | =Further Reading= | ||
| + | |||
| + | * [https://source.ggy.bris.ac.uk/wiki/A_Good_Read#Numerical_Methods] Peter J. Olver has made available some excellent notes on the topic of solving PDEs, including a description of the FEM. | ||
Latest revision as of 17:30, 8 November 2011
Numerical Methods for PDEs: Solving PDEs on a computer
Introduction
Theory
In this tutorial we're going to look at how to solve Partial Differential Equations (PDEs) numerically. That is, on a computer, as opposed to analytically, as we would do using pencil and paper. PDEs crop up a good deal when we describe things in the natural world--e.g. heat flow, or the distribution of electrical charge--and so methods for their solution prove to be widely useful when writing scientific models.
Given the relatively large amount of material that we're about to cover, let's list the main take-home messages straight away:
- Finding a numerical solution to a PDE often boils down (after discritisation) to solving a system of linear equations, of the (matrix) form Ax=b.
- Third-party linear algebra libraries will be much faster and less error prone than your own code for this task. Many of these libraries will also run in parallel.
- By writing code at and above the level of a call to a linear algebra library, we can focus on the higher-level task of doing the science.
- If the matrix A is sparse then iterative methods will be much faster than performing a direct method for solution (such as an LU decomposition).
Practice
In this tutorial we'll take a look at a number of example programs which solve equations using linear algebra packages. These packages include LAPACK, PETSc, PLASMA and MAGMA. The examples will be based upon the solution of a PDE using the Finite Difference Method (FDM), although we'll also touch upon the theory for an alternative approach--the Finite Element Method (FEM).
To download the code for the examples, type:
svn co https://svn.ggy.bris.ac.uk/subversion-open/num-methods1 ./num-methods1
LAPACK (which stands for Linear Algebra PACKage), is the oldest of the packages considered. It has a venerable history and is still highly relevant for some problems today. The project website--which contains download instructions and a great deal of useful documentation--is: http://www.netlib.org/lapack. LAPACK is built on top of the Basic Linear Algebra Subprograms (BLAS) (http://www.netlib.org/blas), but provides a higher level functionality.
Before looking at our first example, we'll start building a 'reference' implementation of LAPACK, as it will take several minutes. First change directory to the location of the LAPACK code that has been bundled into the repository of examples:
cd num-methods1/lapack-3.3.1
and build the libraries, by typing:
make lib
Theory: Looking at Nature (& a quick philosophical aside)
When we observe nature, certain patterns crop up again and again. For example, in the the field of electrostatics, if a function [math]\displaystyle{ f }[/math] describes a distribution of electric charge, then Poisson's equation:
- [math]\displaystyle{ {\nabla}^2 u = f. }[/math]
gives the electric potential [math]\displaystyle{ u }[/math].
However, Poisson's equation also describes the steady state temperature of a material when subjected to some heating and also crops up when considering gravitational potentials..
What's going on here? Why are we using the same equation? Is Poisson's equation fundamental in some way? Well, yes I suppose it is. However, we can look at it another way and say that Poisson's equation is the way we describe--using the language of maths--steady state phenomena which involve potentials. OK, this sounds a bit out there, but consider the following very general equation for a moment (it's a second-order linear equation in two variables):
[math]\displaystyle{ Au_{xx} + Bu_{xy} + Cu_{yy} + Du_x + Eu_y + Fu = G }[/math]
It turns out that we can categorise certain instances of this equation and then relate these categories to the kind of phenomena that they describe:
- Parabolic: [math]\displaystyle{ B^2 - 4AC = 0 }[/math]. This family of equations describe heat flow and diffusion processes.
- Hyperbolic: [math]\displaystyle{ B^2 - 4AC \gt 0 }[/math]. Describe vibrating systems and wave motion.
- Elliptic: [math]\displaystyle{ B^2 - 4AC \lt 0 }[/math]. Steady-state phenomena.
That's pretty handy! OK, we've had our philosophical aside, on with the show.
Practice: Guassian Elimination
While the libraries are building, consider the following system of linear equations:
- [math]\displaystyle{ \begin{alignat}{7} x &&\; + \;&& 3y &&\; - \;&& 2z &&\; = \;&& 5 & \\ 3x &&\; + \;&& 5y &&\; + \;&& 6z &&\; = \;&& 7 & \\ 2x &&\; + \;&& 4y &&\; + \;&& 3z &&\; = \;&& 8 & \end{alignat} }[/math]
(From http://en.wikipedia.org/wiki/System_of_linear_equations.)
which we can write in matrix form:
- [math]\displaystyle{ \begin{bmatrix}1 & 3 & -2\\3 & 5 & 6\\2 & 4 & 3\end{bmatrix} \begin{bmatrix}x\\y\\z\end{bmatrix} = \begin{bmatrix}5\\7\\8\end{bmatrix} }[/math]
The following computation shows Gauss-Jordan elimination applied a further simplified (assumed the x, y and z) matrix version (Ax=b) of the above system, where the operations include:
- top row x3, subtract from 2nd row
- top row x2, subtract from 3rd row
- dived the 2nd row by 4
- multiply the 2nd row by 2, add to the 3rd row
etc:
- [math]\displaystyle{ \left[\begin{array}{rrr|r} 1 & 3 & -2 & 5 \\ 3 & 5 & 6 & 7 \\ 2 & 4 & 3 & 8 \end{array}\right] }[/math][math]\displaystyle{ \sim \left[\begin{array}{rrr|r} 1 & 3 & -2 & 5 \\ 0 & -4 & 12 & -8 \\ 2 & 4 & 3 & 8 \end{array}\right] }[/math][math]\displaystyle{ \sim \left[\begin{array}{rrr|r} 1 & 3 & -2 & 5 \\ 0 & -4 & 12 & -8 \\ 0 & -2 & 7 & -2 \end{array}\right] }[/math][math]\displaystyle{ \sim \left[\begin{array}{rrr|r} 1 & 3 & -2 & 5 \\ 0 & 1 & -3 & 2 \\ 0 & -2 & 7 & -2 \end{array}\right] }[/math][math]\displaystyle{ \sim \left[\begin{array}{rrr|r} 1 & 3 & -2 & 5 \\ 0 & 1 & -3 & 2 \\ 0 & 0 & 1 & 2 \end{array}\right] }[/math][math]\displaystyle{ \sim \left[\begin{array}{rrr|r} 1 & 3 & -2 & 5 \\ 0 & 1 & 0 & 8 \\ 0 & 0 & 1 & 2 \end{array}\right] }[/math][math]\displaystyle{ \sim \left[\begin{array}{rrr|r} 1 & 3 & 0 & 9 \\ 0 & 1 & 0 & 8 \\ 0 & 0 & 1 & 2 \end{array}\right] }[/math][math]\displaystyle{ \sim \left[\begin{array}{rrr|r} 1 & 0 & 0 & -15 \\ 0 & 1 & 0 & 8 \\ 0 & 0 & 1 & 2 \end{array}\right]. }[/math]
OK, so far so good? Let's try our first code example:
cd ~/num-methods1/examples/example1
To see how to solve the above matrix example using LAPACK take a look inside dgesv-example.f90. First the declarations:
integer, parameter :: N = 3
integer, parameter :: LDA = N ! leading dimension of A
integer, parameter :: LDB = N ! leading dimension of B
integer, parameter :: NRHS = 1 ! no. of RHS, i.e columns in b
integer, dimension(N) :: IPIV
integer :: INFO
integer :: i
real(kind=8), dimension(LDA,N) :: A ! LDAxN matrix
real(kind=8), dimension(LDB,NRHS) :: B ! LDBxNRHS matrix
Where, we see that the leading dimension of a matrix is used to specify the number of rows of storage. Note also, that we could simultaneously solve for multiple vectors B, by specifying NRHS > 1.
Next, we set up the matrix A:
! ( 1 3 -2)
! ( 3 5 6)
! ( 2 4 3)
A(1,1) = 1.0d+0
A(1,2) = 3.0d+0
A(1,3) = -2.0d+0
A(2,1) = 3.0d+0
A(2,2) = 5.0d+0
A(2,3) = 6.0d+0
A(3,1) = 2.0d+0
A(3,2) = 4.0d+0
A(3,3) = 3.0d+0
And the vector B:
! ( 5)
! ( 7)
! ( 8)
B(1,1) = 5.0d+0
B(2,1) = 7.0d+0
B(3,1) = 8.0d+0
Once we have all that in place, all we need to do is call the solver routine:
call dgesv(N, NRHS, A, LDA, IPIV, B, LDB, INFO)
Note that the solution vector replaces the values in B. (The contents of A will be changed in place too, but more of that later.)
! Print the result vector X.
! It should be:
! (-15)
! ( 8)
! ( 2)
do ii=1,LDB
write(*,*) B(ii,1)
end do
Run the program to satisfy yourself that it works:
cd num-methods1/examples/example1 make ./dgesv-example.exe
The Finite Difference Method
In this section we'll look at solving PDEs using the Finite Difference Method. Finite difference equations have a long and illustrious past. Indeed, they were used by Newton-and-the-gang as a logical route in to talk about differential calculus. So, in fact, we're just going to work back in the other direction..
Theory: Discritisation
The functions and variables that we see in, for instance, Poisson's equation are continuous; meaning that they could have an infinite number of values. This is fine when solving equations analytically, but we are limited to considering only a finite number of values when solving PDEs using a digital computer. We need a way to discritise our problems. Happily, help is at hand from the (sometimes tortuous) Taylor series: We can approximate the value of a function at a point distant from x (i.e. displaced by an amount h) using a linear summation of derivatives of the function at x--a Taylor expansion:
- [math]\displaystyle{ f(x+h) \approx f(x) + \frac{f'(x)}{1!}h + \frac{f''(x)}{2!}h^2 + \cdots }[/math]
We can truncate this series after the first differential and rearrange, to get ourselves the forward-difference approximation:
- [math]\displaystyle{ f'(x) \approx \frac{f(x+h) - f(x)}{h} }[/math]
Thus, following the finite difference approach, we can discritise our equations by replacing our differential operators by their difference approximations, as helpfully provided by a truncated Taylor series. If, like me, you like thinking visually, the schematic below shows the relationship between a differential operator--the gradient at a point [math]\displaystyle{ x_1 }[/math] and the finite difference approximation to the gradient:
If we were to consider a displacement of -h instead, we would get the backward-difference approximation to [math]\displaystyle{ f'(x) }[/math]:
- [math]\displaystyle{ f'(x) \approx \frac{f(x) - f(x-h)}{h} }[/math],
Subtracting the backward difference from the forward difference we get the central-difference approximation:
- [math]\displaystyle{ f'(x) \approx \frac{1}{2h}(f(x+h) - f(x-h)) }[/math].
By retaining an extra term from the Taylor series, and some similar rearrangements, we can also formulate a finite difference approximation to the 2nd differential. For example the central-difference formula for this is:
- [math]\displaystyle{ f''(x) \approx \frac{1}{h^2} [ f(x+h) - 2f(x) + f(x-h) ] }[/math].
In a similar way we can derive formulae for approximations to partial derivatives. Armed with all these formulae, we can convert differential equations into difference equations, which we can solve on a computer. However, since we are dealing with approximations to the original equation(s), we must be vigilant that errors in our input and intermediate calculations do not accumulate such that our final solution is meaningless.
Theory: Eliptical Equations & the Discrete Laplacian Operator
Now that we have our difference formulae, let's apply them to Laplace's equation in 2-dimensions:
- [math]\displaystyle{ \frac{\partial^2u}{\partial x^2} + \frac{\partial^2u}{\partial y^2} = 0 }[/math]
This describes purely diffusive phenomena and can give us, for example, the steady state temperature over a 2D plate. Let's suppose that three sides of the plate are held at 100°C, while the fourth is held at 0°C. We can divide the plate into a number of grid cells:
Using the central difference approximation to a 2nd differential that we derived in the last section, we can rewrite the differential equation as a difference equation:
- [math]\displaystyle{ \frac{\partial^2u}{\partial x^2} + \frac{\partial^2u}{\partial y^2} \approx \frac{1}{h^2}(u^j_{i+1} -2u^j_i +u^j_{i-1}) + \frac{1}{k^2}(u^{j+1}_i -2u^j_i +u^{j-1}_i) }[/math]
where h is the grid spacing in the i-direction and k is the spacing the j-direction. If we set [math]\displaystyle{ h=k }[/math], then:
- [math]\displaystyle{ u^j_{i+1} + u^j_{i-1} + u^{j+1}_i + u^{j-1}_i - 4u^j_i = 0 }[/math]
solving for [math]\displaystyle{ u^j_i }[/math]:
- [math]\displaystyle{ u^j_i = \frac{1}{4}(u^j_{i+1} + u^j_{i-1} + u^{j+1}_i + u^{j-1}_i) }[/math]
Note that this last equation says--quite intuitively--that the value at the central grid point is equal to the average of its four neighbours. We can draw a picture of the stencil needed to compute the central value:
Centring the stencil on each of the 4 grid cells of the inner region (orange) in turn, we generate the following 4 difference equations:
- [math]\displaystyle{ \begin{alignat}{5} -4u_{22} & + u_{32} & + u_{23} & + 100 & + 100 & = 0 \\ -4u_{32} & + 0 & + u_{33} & + u_{22} & + 100 & = 0 \\ -4u_{23} & + u_{33} & + 100 & + 100 & + u_{22} & = 0 \\ -4u_{33} & + 0 & + 100 & + u_{23} & + u_{32} & = 0 \\ \end{alignat} }[/math]
This system of equations can be written in matrix form:
- [math]\displaystyle{ \begin{bmatrix}-4 & 1 & 1 & 0\\1 & -4 & 0 & 1\\1 & 0 & -4 & 1\\0 & 1 & 1 & -4\end{bmatrix} \begin{bmatrix}u_{22}\\u_{32}\\u_{23}\\u_{33}\end{bmatrix} = \begin{bmatrix}-200\\-100\\-200\\-100\end{bmatrix} }[/math]
Which may be summarised as the vector equation, Ax=b, which can be efficiently & conveniently solved using a library linear algebra routines, such as LAPACK, PETSc or PLASMA.
Practice: Solving Eliptical Problems using LAPACK
Next, take a look at simple-laplace.f90. This program solves the 2D laplace equation for a 4x4 domain, as described above. This program only differs from the previous one in way in which we initialise the matrix A and vector b. To run the program type:
./simple-laplace.exe
mxm-laplace.f90 also solves our laplacian example, except that this time we can vary the size of the matrix and corresponding arrays for an mxm square domain:
./mxm-laplace.exe
LAPACK also gives us access to a routine--dgesvx--which solves the same system of equations, but also gives us some error analysis. The x stands for expert.
./mxm-laplace-expert.exe
Exercises
- Play around with mxm-laplace.f90. Try different domain sizes, and different boundary conditions.
Practice: Details of the LU Decomposition
Actually, the LAPACK routine dgesv does not perform Gaussian elimination, but rather computes an LU decomposition and then solves the equation using two substitution steps--one forward and one backward. This is because for the same computational cost, we can factor A without knowing about b. We can then find x cheaply, for many different b. This is useful when solving parabolic equations, where x at time-step [math]\displaystyle{ j }[/math] forms the basis for b at time-step [math]\displaystyle{ j+1 }[/math].
OK, how to we obtain an LU decomposition?
The Guassian elimination process above can be expressed as a matrix multiplication. The elimination in the first column can be achieved by left multiplying A with a unit (1's on the diagonal) lower triangular matrix, [math]\displaystyle{ L_1 }[/math], with non-zeros only in the first column (this case is called an atomic unit lower triangular matrix), such as that schematically shown below:
- [math]\displaystyle{ \begin{bmatrix} 1 & 0 & 0 \\ x & 1 & 0 \\ y & 0 & 1 \end{bmatrix} }[/math]
Following our example, [math]\displaystyle{ L_1 A=A' }[/math] becomes:
- [math]\displaystyle{ \begin{bmatrix} 1 & 0 & 0 \\ -3 & 1 & 0 \\ -2 & 0 & 1 \end{bmatrix} \times \begin{bmatrix} 1 & 3 & -2 \\ 3 & 5 & 6 \\ 2 & 4 & 3 \end{bmatrix} = \begin{bmatrix} 1 & 3 & -2 \\ 0 & -4 & 12 \\ 0 & -2 & 7 \end{bmatrix} }[/math]
The eliminations in the second column are achieved by left multiplying [math]\displaystyle{ A'=L_1 A }[/math] by [math]\displaystyle{ L_2 }[/math] (which is atomic unit lower triangular with non-zeros only in the second column):
- [math]\displaystyle{ \begin{bmatrix} 1 & 0 & 0 \\ 0 & 1 & 0 \\ 0 & -\frac{1}{2} & 1 \end{bmatrix} \times \begin{bmatrix} 1 & 0 & 0 \\ -3 & 1 & 0 \\ -2 & 0 & 1 \end{bmatrix} \times \begin{bmatrix} 1 & 3 & -2 \\ 3 & 5 & 6 \\ 2 & 4 & 3 \end{bmatrix} = \begin{bmatrix} 1 & 3 & -2 \\ 0 & -4 & 12 \\ 0 & 0 & 1 \end{bmatrix} }[/math]
We note that the result [math]\displaystyle{ L_2 L_1 A = A'' = U }[/math] is upper triangular. Rearranging we get, [math]\displaystyle{ A=L_1^{-1} \cdots L^{-1}_{n-2} L^{-1}_{n-1} U }[/math]. The inverse matrices, [math]\displaystyle{ L^{-1} }[/math], are very easy to find as the inverse of an atomic unit lower triangular matrix is the same as the original save only for a change of sign in the off diagonal elements. The product is, again, unit lower triangular. Thus [math]\displaystyle{ A=LU }[/math]:
- [math]\displaystyle{ \begin{bmatrix} 1 & 3 & -2 \\ 3 & 5 & 6 \\ 2 & 4 & 3 \end{bmatrix} = \begin{bmatrix} 1 & 0 & 0 \\ 0 & 1 & 0 \\ 0 & \frac{1}{2} & 1 \end{bmatrix} \times \begin{bmatrix} 1 & 0 & 0 \\ 3 & 1 & 0 \\ 2 & 0 & 1 \end{bmatrix} \times \begin{bmatrix} 1 & 3 & -2 \\ 0 & -4 & 12 \\ 0 & 0 & 1 \end{bmatrix} = \begin{bmatrix} 1 & 0 & 0 \\ 3 & 1 & 0 \\ 2 & \frac{1}{2} & 1 \end{bmatrix} \times \begin{bmatrix} 1 & 3 & -2 \\ 0 & -4 & 12 \\ 0 & 0 & 1 \end{bmatrix} }[/math]
Given L and U we can solve Ax=b by first solving Ly=b, where:
- [math]\displaystyle{ \begin{bmatrix} 1 & 0 & 0 \\ 3 & 1 & 0 \\ 2 & \frac{1}{2} & 1 \end{bmatrix} \begin{bmatrix}y_1\\y_2\\y_3\end{bmatrix} = \begin{bmatrix}5\\7\\8\end{bmatrix} }[/math]
As L is unit lower triangular, we can do this by easily by forward substitution:
- [math]\displaystyle{ \begin{array}{rcl} y_1 & = & 5 \\ 3 \times y_1 + y_2 & = & 7 \\ y_2 & = & 7 - 15 \\ y_2 & = & -8 \\ 10 - 4 + y_3 & = & 8 \\ y_3 & = & 12 - 10 \\ y_3 & = & 2 \end{array} }[/math]
Once we know y, we can solve Ux=y:
- [math]\displaystyle{ \begin{bmatrix} 1 & 3 & -2 \\ 0 & -4 & 12 \\ 0 & 0 & 1 \end{bmatrix} \begin{bmatrix}x_1\\x_2\\x_3\end{bmatrix} = \begin{bmatrix}5\\-8\\2\end{bmatrix} }[/math]
This time easily by backward substitution:
- [math]\displaystyle{ \begin{array}{rcl} x_3 & = & 2 \\ 24 - 4 \times x_2 & = & -8 \\ 32 & = & 4x_2 \\ x_2 & = & 8 \\ -4 + 24 + x_1 & = & 5 \\ x_1 & = & 9 - 24 \\ x_1 & = & -15 \end{array} }[/math]
Computing the LU decomposition of an [math]\displaystyle{ n \times n }[/math] system of equations has a computational cost of [math]\displaystyle{ O(n^3) }[/math] operations, whereas the forward and backward substitution steps have a cost of [math]\displaystyle{ O(n^2) }[/math] operations. Thus the cost of solution is dominated by the LU decomposition.
To demonstrate that the solution does indeed take [math]\displaystyle{ O(n^3) }[/math] operations, I ran mxm-laplacian.exe on my desktop PC for varying sizes of N and could plot the following from the results:
If we look once again at dgesv-example.f90 and set:
logical, parameter :: verbose=.true.
(and recompile and run) we can see the contents of A after the call to dgesv(). It now contains an encoding of the LU decomposition (the unit leading diagonal from L is assumed):
LU decomposition as encoded in matrix A:
3.00000000000000 5.00000000000000 6.00000000000000
0.333333333333333 1.33333333333333 -4.00000000000000
0.666666666666667 0.500000000000000 1.00000000000000
..and the IPIV vector:
2 2 3
This is all well and good, but hang-on a cotton pickin' minute!, the matrix U is not as we expected:
- [math]\displaystyle{ \begin{bmatrix} 3 & 5 & 6 \\ 0 & \frac{4}{3} & -4 \\ 0 & 0 & 1 \end{bmatrix} \neq \begin{bmatrix} 1 & 3 & -2 \\ 0 & -4 & 12 \\ 0 & 0 & 1 \end{bmatrix} }[/math]
Why not? The answer lies the in contents of the IPIV. The elements along the leading diagonal are known as pivots. If any pivots are small, our algorithm will become unstable. We may exchange rows (known as partial pivoting) in A without changing the solution. We can read the elements of IPIV as a set of instructions for manipulating each row of the original matrix.
For example, if the first element of IPIV is a 1, then we read this as:
- exchange row 1 with row 1 (i.e. no change--a null operation)
but if the first element of IPIV were a 2, then this means:
- exchange row 1 with row 2 (i.e. swap round the firs two rows)
In the case of our example program, the following sequence of actions were performed:
- row 1 was exchanged with row 2
- row 2 was exchanged with row 2
- row 3 was exchanged with row 3
We can satisfy ourselves that all is well, by factorising this permuted matrix:
- [math]\displaystyle{ \begin{bmatrix} 1 & 0 & 0 \\ 0 & 1 & 0 \\ 0 & \frac{1}{2} & 1 \end{bmatrix} \times \begin{bmatrix} 1 & 0 & 0 \\ \frac{1}{3} & 1 & 0 \\ \frac{2}{3} & 0 & 1 \end{bmatrix} \times \begin{bmatrix} 3 & 5 & 6 \\ 0 & \frac{4}{3} & -4 \\ 0 & 0 & 1 \end{bmatrix} = \begin{bmatrix} 3 & 5 & 6 \\ 1 & 3 & -2 \\ 2 & 4 & 3 \end{bmatrix} }[/math]
Theory: Parabolic Equations & Forward (Explicit) Euler
Let's look at how to solve a different class of equation, again using finite difference techniques.
An example of a parabolic equation is the 1D heat equation, which describes the evolution of temperature at various points in a thin, insulated bar over time. Imagine such a bar where the ends are kept at 0°C:
Since this scenario involves the passage of time, just stating the boundary conditions is not enough, and we must also specify some initial condition for the temperature along the bar's length. This is shown below for a bar divided up into grid cells, as before:
The 1D heat equation is:
- [math]\displaystyle{ \frac{\partial u}{\partial t}=\alpha \frac{\partial^2u}{\partial x^2} }[/math]
where [math]\displaystyle{ \alpha }[/math]--the thermal diffusivity--is set to 1 for simplicity. One way to rewrite this as a difference equation (using a forward difference for the LHS and a central difference for the RHS*) is:
- [math]\displaystyle{ \frac{u^{j+1}_i - u^j_i }{k} = \frac{u^j_{i+1} - 2u^j_i + u^j_{i-1}}{h^2} }[/math]
and rearrange to solve for [math]\displaystyle{ u^{j+1}_i }[/math]:
- [math]\displaystyle{ u^{j+1}_i = u^j_i + r(u^j_{i+1} - 2u^j_i + u^j_{i-1}) }[/math]
- [math]\displaystyle{ u^{j+1}_i = ru^j_{i+1} + (1- 2r)u^j_i + ru^j_{i-1} }[/math]
where:
[math]\displaystyle{ r=\frac{k}{h^2} }[/math]
This time, we see that the appropriate stencil is:
If we set [math]\displaystyle{ h=\frac{1}{10} }[/math] and [math]\displaystyle{ k=\frac{1}{1000} }[/math], then [math]\displaystyle{ r=\frac{1}{10} }[/math], and the equation becomes:
- [math]\displaystyle{ u^{j+1}_i = u^j_i + \frac{1}{10}(u^j_{i+1} - 2u^j_i + u^j_{i-1}) }[/math]
- [math]\displaystyle{ u^{j+1}_i = \frac{1}{10}(u^j_{i+1} + 10u^j_i - 2u^j_i + u^j_{i-1}) }[/math]
- [math]\displaystyle{ u^{j+1}_i = \frac{1}{10}(u^j_{i+1} + 8u^j_i + u^j_{i-1}) }[/math]
Which we can illustrate using weightings on the stencil nodes:
Forward Euler is as an explicit method, as one unknown value ([math]\displaystyle{ u^{j+1}_i }[/math]) is expressed directly in terms of known values (all values of u at time j are known).
We could implement a simple marching algorithm, where we centre the stencil over a set of known values at time j, and may calculate the unknown temperature for grid cell i at time j+1. However, it would be better to group together all the calculations for a particular time step into a system of simultaneous equations (for our example 1d heat problem):
- [math]\displaystyle{ u_{22} = \frac{1}{10}(0) + \frac{8}{10}(0.2) + \frac{1}{10}(0.4) }[/math]
- [math]\displaystyle{ u_{23} = \frac{1}{10}(0.2) + \frac{8}{10}(0.4) + \frac{1}{10}(0.6) }[/math]
- [math]\displaystyle{ u_{24} = \frac{1}{10}(0.4) + \frac{8}{10}(0.6) + \frac{1}{10}(0.8) }[/math]
- [math]\displaystyle{ \cdots }[/math]
and then re-write in matrix form:
- [math]\displaystyle{ \begin{bmatrix}u_{22} \\ u_{23} \\ u_{24} \\\vdots \end{bmatrix} = \begin{bmatrix} 0.1 & 0.8 & 0.1 & 0 & 0 & \cdots & 0 \\ 0 & 0.1 & 0.8 & 0.1 & 0 & \cdots & 0 \\ \vdots & & & & & & \vdots \\ & & \cdots & 0 & 0.1 & 0.8 & 0.1 \\ \end{bmatrix} \times \begin{bmatrix}0 \\ 0.2 \\ 0.4 \\ \vdots \\ 0.2 \\ 0 \end{bmatrix} }[/math]
as the computational efficiency of a matrix vector multiplication will be hard to better. Note that we can write the above piece of linear algebra in a general form:
- [math]\displaystyle{ \begin{bmatrix}u_{22} \\ u_{23} \\ u_{24} \\ \vdots \end{bmatrix} = \begin{bmatrix} r & 1-2r & r & 0 & 0 & \cdots & 0 \\ 0 & r & 1-2r & r & 0 & \cdots & 0 \\ \vdots & & & & & & \vdots \\ & & \cdots & 0 & r & 1-2r & r \\ \end{bmatrix} \times \begin{bmatrix}u_{11} \\ u_{12} \\ u_{13} \\ u_{14} \\ \vdots \end{bmatrix} }[/math]
(*The two difference schemes for either side of the equation were chosen so that the approximation error characteristics for either side are approximately balanced and so the errors on one side or the other do not begin to dominate the calculations.)
Theory: Solution Stability and the CFL Condition
The Forward Euler scheme has the benefit of simplicity, but is unstable for large time steps. The CFL (Courant–Friedrichs–Lewy) condition states that the following must hold:
- [math]\displaystyle{ r=\frac{k}{h^2} \le 0.5 }[/math]
for stability.
Theory: Parabolic Equations & Backward (Implicit) Euler
Another way to formulate our finite difference equations for the very same problem is to use backward-differences instead of forward-differences. The backward Euler scheme requires more computation per time-step than the forward scheme, but has the distinct advantage that it is unconditionally stable with regard to the relative size between the time and spatial step sizes; i.e. we are not limited to [math]\displaystyle{ r \le 0.5 }[/math] when we use the backward Euler scheme.
We will use a central-difference on the RHS, as before, but shifting the time index to [math]\displaystyle{ j+1 }[/math]. This will allow us to formulate a backward-difference on the RHS between the time indices [math]\displaystyle{ j }[/math] and [math]\displaystyle{ j+1 }[/math]:
- [math]\displaystyle{ \frac{u^{j+1}_i - u^j_i }{k} = \frac{u^{j+1}_{i+1} - 2u^{j+1}_i + u^{j+1}_{i-1}}{h^2} }[/math]
- [math]\displaystyle{ u^{j+1}_i - u^j_i = \frac{k}{h^2}(u^{j+1}_{i+1} - 2u^{j+1}_i + u^{j+1}_{i-1}) }[/math]
- [math]\displaystyle{ u^{j+1}_i - u^j_i = r(u^{j+1}_{i+1} - 2u^{j+1}_i + u^{j+1}_{i-1}) }[/math]
- [math]\displaystyle{ u^{j+1}_i - u^j_i = ru^{j+1}_{i+1} - 2ru^{j+1}_i + ru^{j+1}_{i-1} }[/math]
- [math]\displaystyle{ u^{j+1}_i - ru^{j+1}_{i+1} + 2ru^{j+1}_i - ru^{j+1}_{i-1} = u^j_i }[/math]
- [math]\displaystyle{ u^j_i = - ru^{j+1}_{i+1} + (1 + 2r)u^{j+1}_i - ru^{j+1}_{i-1} }[/math]
If we again choose [math]\displaystyle{ h=\frac{1}{10} }[/math] and [math]\displaystyle{ k=\frac{1}{1000} }[/math], such that [math]\displaystyle{ r=\frac{k}{h^2}=\frac{1}{10} }[/math], as before, we end up with the following system of equations when we consider all values of [math]\displaystyle{ i }[/math] for our first time-step--from [math]\displaystyle{ j }[/math] to [math]\displaystyle{ j+1 }[/math]--where we know values of all the cells at time [math]\displaystyle{ j }[/math]:
- [math]\displaystyle{ 0.2 = -\frac{1}{10}(0) + \frac{12}{10}(u_{22}) -\frac{1}{10}(u_{23}) }[/math]
- [math]\displaystyle{ 0.4 = -\frac{1}{10}(u_{22}) + \frac{12}{10}(u_{23}) -\frac{1}{10}(u_{24}) }[/math]
- [math]\displaystyle{ 0.6 = -\frac{1}{10}(u_{23}) + \frac{12}{10}(u_{24}) -\frac{1}{10}(u_{25}) }[/math]
- [math]\displaystyle{ \cdots }[/math]
We can see that this is akin to looping over the domain with the following stencil:
Rearranging, so that we have only constants on the RHS:
- [math]\displaystyle{ \frac{1}{10}(0) + \frac{12}{10}(u_{22}) -\frac{1}{10}(u_{23}) = 0.2 + \frac{1}{10}(0) }[/math]
- [math]\displaystyle{ \frac{1}{10}(u_{22}) + \frac{12}{10}(u_{23}) -\frac{1}{10}(u_{24}) = 0.4 }[/math]
- [math]\displaystyle{ \frac{1}{10}(u_{23}) + \frac{12}{10}(u_{24}) -\frac{1}{10}(u_{25}) = 0.6 }[/math]
- [math]\displaystyle{ \cdots }[/math]
We can re-write this system of equations in matrix form:
- [math]\displaystyle{ \begin{bmatrix} 1.2 & -0.1 & 0 & 0 & \cdots & 0 \\ -0.1 & 1.2 & -0.1 & 0 & \cdots & 0 \\ \vdots & & & & & \vdots \\ & & \cdots & 0 & -0.1 & 1.2 \\ \end{bmatrix} \times \begin{bmatrix} u_{22} \\ u_{23} \\ u_{24} \\\vdots \end{bmatrix} = \begin{bmatrix} 0.2 + 0.1(0) \\ 0.4 \\ \vdots \\ 0.2 +0.1(0) \end{bmatrix} }[/math]
The general form is:
- [math]\displaystyle{ \begin{bmatrix} 1+2r & -r & 0 & 0 & \cdots & 0 \\ -r & 1+2r & -r & 0 & \cdots & 0 \\ \vdots & & & & & \vdots \\ & & \cdots & 0 & -r & 1+2r \\ \end{bmatrix} \times \begin{bmatrix} u_{22} \\ u_{23} \\ u_{24} \\\vdots \end{bmatrix} = \begin{bmatrix} u_{12} + r(u_{11}) \\ u_{13} \\ \vdots \\ u_{1n-1} + r(u_{1n}) \end{bmatrix} }[/math]
Theory: Parabolic Equations & Crank-Nicolson
Now, forward and backward Euler schemes are by far from the only ways in which to approach this 1D heat problem. Yet another scheme is the to create finite difference equations using the Crank-Nicolson stencil:
The stencil amounts to an average of both the forward and backward Euler methods. The resulting difference equation is:
- [math]\displaystyle{ \frac{u^{j+1}_i - u^j_i}{k} = \frac{1}{2}(\frac{u^{j+1}_{i+1} - 2u^{j+1}_{i} + u^{j+1}_{i-1}}{h^2} + \frac{u^j_{i+1} - 2u^j_i + u^j_{i-1}}{h^2}). }[/math]
Which we may rearrange to become:
- [math]\displaystyle{ u^{j+1}_i - u^j_i = \frac{1}{2}\frac{k}{h^2}(u^{j+1}_{i+1} - 2u^{j+1}_i + u^{j+1}_{i-1} + u^j_{i+1} - 2u^j_i + u^j_{i-1}) }[/math]
- [math]\displaystyle{ 2(u^{j+1}_i - u^j_i) = r(u^{j+1}_{i+1} - 2u^{j+1}_i + u^{j+1}_{i-1} + u^j_{i+1} - 2u^j_i + u^j_{i-1}) }[/math],
where [math]\displaystyle{ r=\frac{k}{h^2} }[/math].
Collecting the unknowns (values of u at the next timestep, [math]\displaystyle{ j+1 }[/math]) to the LHS and the knowns (values of u at current timestep, [math]\displaystyle{ j }[/math]), we get:
- [math]\displaystyle{ 2u^{j+1}_i + 2ru^{j+1}_i - ru^{j+1}_{i+1} - ru^{j+1}_{i-1} = 2u^j_i +ru^j_{i+1} -2ru^j_i + ru^j_{i-1} }[/math]
- [math]\displaystyle{ (2+2r)u^{j+1}_i - ru^{j+1}_{i+1} - ru^{j+1}_{i-1} = (2-2r)u^j_i +ru^j_{i+1} + ru^j_{i-1} }[/math].
If we choose [math]\displaystyle{ h=1/10 }[/math] and [math]\displaystyle{ k=1/100 }[/math] (note that the time-step is 10x larger than than chosen for the forward Euler example), then [math]\displaystyle{ r=1 }[/math], and we get:
- [math]\displaystyle{ -u_{i-1}^{j+1} +4u_i^{j+1} -u_{i+1}^{j+1} = u_{i-1}^j + u_{i+1}^j }[/math].
We can again create a system of linear equations for all the cells j along the bar at a given time step i:
- [math]\displaystyle{ -0 + 4u_{22} - u_{23} = 0 + 0.4 }[/math]
- [math]\displaystyle{ -u_{22} + 4u_{23} - u_{24} = 0.2 + 0.6 }[/math]
- [math]\displaystyle{ -u_{23} + 4u_{24} - u_{25} = 0.4 + 0.8 }[/math]
- [math]\displaystyle{ \cdots }[/math]
We have [math]\displaystyle{ j }[/math] unknowns and [math]\displaystyle{ j }[/math] equations. Then we can again translate into matrix notation and solve using a library linear algebra routines:
- [math]\displaystyle{ \begin{bmatrix} -4 & 1 & 0 & 1 & 0 & 0 & \cdots & 0 \\ 1 & -4 & 1 & 0 & 1 & 0 & \cdots & 0 \\ \vdots & & & & & & & \vdots \\ & \cdots & & 0 & 1 & 0 & 1 & -4\\ \end{bmatrix} \times \begin{bmatrix} u_{22} \\ u_{23} \\ u_{24} \\\vdots \end{bmatrix} = \begin{bmatrix} 0 + 0.4 \\ 0.2 + 0.6 \\ \vdots \\ 0 + 0.4 \end{bmatrix} }[/math]
In general:
- [math]\displaystyle{ \begin{bmatrix} -(2+2r) & r & 0 & r & 0 & 0 & \cdots & 0 \\ r & -(2+2r) & r & 0 & r & 0 & \cdots & 0 \\ \vdots & & & & & & & \vdots \\ & \cdots & & 0 & r & 0 & r & -(2+2r)\\ \end{bmatrix} \times \begin{bmatrix} u_{22} \\ u_{23} \\ u_{24} \\\vdots \end{bmatrix} = \begin{bmatrix} ru_{11} + (2-2r)u_{12} + ru_{13} \\ \vdots \end{bmatrix} }[/math]
Practice: Solving Parabolic Problems using LAPACK
In our second example, we'll see 3 different schemes for solving a parabolic problem--forward Euler, backward Euler and Crank-Nicolson. The forward Euler scheme is explicit and so no system of linear equations is solved. It is a appealingly simple scheme, but can suffer from unstable solutions under certain conditions. In both the backward Euler and Crank-Nicolson schemes, we will see the (repeated) use of a pre-computed LU decomposition when solving a parabolic equation.
cd ../example2
In simple-crank-nicolson.f90 we see that the initial call to dgesv replaces the entries in the matrix A with an encoding of the LU decompostion (the unit diagonal is assumed). For subsequent timesteps we can thus call dgetrs instead:
do ii=2,4
...
call dgetrs('N', N, NRHS, A, LDA, IPIV, B, LDB, INFO)
...
end do
where we have remembered to take account of the boundary conditions at each time-step too:
do ii=2,4
...
! copy B into B1, so that we can recompute for
! the boundary conditions again.
B1 = B
! Need to recalculate entries for the vector of known
! values--held in B--given the solution at the current
! timestep--held in B1.
! *NB* the loops below are only valid for the case r=1.
! LHS
B(1,1) = 0 + B1(2,1)
! RHS
B(N,1) = 0 + B1(N-1,1)
! middle bit
do jj=2,N-1
B(jj,1) = B1(jj-1,1) + B1(jj+1,1)
end do
...
end do
Exercises
- Experiment with values of [math]\displaystyle{ r=\frac{k}{h^2} }[/math] for the 3 schemes, so as to highlight the effects of the CFL condition.
Practice: Banded Matrices
In all my examples, thus far, I've called routines from the dgexxx series offered by LAPACK. The d stands for double and the ge for generel purpose. For banded matrices, LAPACK offers a much more efficient format and routines to match. Take a look at http://www.netlib.org/lapack/double/ for a list of driver routines using matrices of double precision numbers, or http://www.netlib.org/lapack for more documentation in general.
Practice: PETSc & Iterative solutions and sparse matrices
In order to compute the solution of a linear system via the direct method of an LU-decomposition, we know that it will cost [math]\displaystyle{ O(n^3) }[/math] operations. Can we compute a solution more cheaply? The class of iterative methods strive to compute a cheaper solution by first guessing a value for the solution vector [math]\displaystyle{ x }[/math] and then iteratively refining it. Thus we will get a series of (hopefully!) improving approximations:
- [math]\displaystyle{ x^0, x^1, x^2, x^3, \cdots, x^j \rightarrow x }[/math]
By way of an illustration of such a method, consider splitting matrix A into 3 others:
- [math]\displaystyle{ A = L + D + U }[/math]
Note the summation, and so the L and the U matrices are not the same as those considered in the LU-decomposition used by the direct approach. With this split, Ax=b, becomes:
- [math]\displaystyle{ Lx + Dx + Ux = b }[/math]
The game now is to replace some occurrences of [math]\displaystyle{ x }[/math] by [math]\displaystyle{ x^k }[/math] and others by [math]\displaystyle{ x^{k+1} }[/math], and thereby obtaining a relationship which tells us how to get an approximation at one iteration step, from the approximation at the previous step. If the resulting iterative scheme converges, then we're in business!
One classic form of such an iteration is the Jacobi method (which is useful when A is diagonally dominant):
- [math]\displaystyle{ Lx^j + Dx^{j+1} + Ux^j = b }[/math]
- [math]\displaystyle{ x^{j+1} = D^{-1}(b - (L + U)x^j) }[/math]
To take a look at a illustrative implementation of the Jacobi method using LAPACK:
cd ../example3 make
and take a look at jacobi-example.f90. To run the executable, type:
./jacobi-example.exe
Happily, however, we do not need to write our own iterative schemes, as the good folk at Argonne have written the PETSc library.
http://www.mcs.anl.gov/petsc/petsc-as/
Take a look inside ex1f.F to see how to solve our laplacian example using PETSc. To run the program you will obviously need PETSc installed on the computer you are using. If you have, then edit Makefile and set PETSC_DIR to the appropriate value, and then type:
make ex1f qsub mpi_submit
Look in the file called OUT for the results of running PETSc.
We can again plot a graph of timings given varying N:
We can see that an iterative method can scale approximately [math]\displaystyle{ O(n^2) }[/math]. For large matrices, this can mean that a solution is found several orders of magnitude faster. Now that is a speed-up worth having! It's liking buying a supercomputer than runs several orders of magnitude faster than your old one..
Better yet, you can try different schemes for iterative solution, just by varying some command-line arguments. For example:
./ex1f -ksp_type bcgs -pc_type jacobi
will invoke the biconjugate gradient stabalised scheme together with a jacobi preconditioner matrix. (This in fact runs faster than the GMRES method for the above case--808ms for N=9694. Note, however, that it cannot scale better than [math]\displaystyle{ O(N^2) }[/math].) For those interested, the PETSc manual is the best next port of call for more details on the various methods made available by the package.
For further examples see INSTALL_DIR/petsc-3.1-p1/src/ksp/ksp/examples/tutorials.
Other Numerical Methods
We have looked at the finite difference method (FDM) above, but other (more mathsy) approaches exist, such as the finite element and finite volume methods. Each approach has its pros and cons. For example, the FDM is limited to regular grids spread over rectangular domains, but is easy to program. In contrast the FEM is applicable to intricately shaped domains and the resolution of the individual elements can be varied, but is requires more involved programming and a deeper level of understand of the relevant mathematics. It should also be noted, however, that under certain circumstances, the FEM and FVM boil down to be exactly the same as the FDM, even though we've approached the task of solving a PDE from a different direction.
One thing which is always common to the approaches is that the problem boils down to a system of linear equations to solve, and that a library of linear algebra routines will be the workhorse when finding solutions to PDEs on a computer. Repeat after me, "a fast linear algebra library is key!"
Theory: The Finite Element Method (FEM)
The general approach here is to create a potential function from our PDE and to find a solution to it via minimising the potential function. Key steps include:
- Discritisation of the original PDE, often by proposing that we can approximate the solution using a linear sum of (polynomial) basis functions.
- Create the potential function by invoking the method of weighted residuals, and in particular the Galerkin specialisation of this approach.
- Use whatever algebraic manipulation and integration (possibly numerical) is required to create a system of linear equations.
- Solve said system of linear equations and in doing so, solve the original PDE.
For example, consider again Poisson's equation over a 2-dimensional region, [math]\displaystyle{ R }[/math]:
- [math]\displaystyle{ \nabla^2u=f(x,y) }[/math]
where [math]\displaystyle{ u=0 }[/math] at the boundary of the domain.
Next, applying the method of weighted residuals, we multiply both sides by weighting functions [math]\displaystyle{ \phi_i }[/math], and take integrals over the domain:
- [math]\displaystyle{ \iint\limits_R \nabla^2u\phi_i\, dA = \iint\limits_R f\phi_i\, dA }[/math]
We can apply integration-by-parts to the LHS ([math]\displaystyle{ \phi_i\nabla^2u }[/math]), for example:
- [math]\displaystyle{ \int \phi_i\nabla^2 u = \phi_i\nabla u - \int \nabla\phi_i \cdot \nabla u }[/math]
Differentiating both sides:
- [math]\displaystyle{ \phi_i\nabla^2 u = \nabla \cdot (\phi_i\nabla u) - \nabla\phi_i \cdot \nabla u }[/math]
So inserting this into our equation for weighted residuals, above:
- [math]\displaystyle{ \iint\limits_R [\nabla \cdot (\phi_i\nabla u) - \nabla\phi_i \cdot \nabla u]\, dA = \iint\limits_R f\phi_i\, dA }[/math]
and rearranging, we get:
- [math]\displaystyle{ \iint\limits_R \nabla\phi_i \cdot \nabla u\, dA = \iint\limits_R \nabla \cdot (\phi_i\nabla u)\, dA - \iint\limits_R f\phi_i\, dA }[/math]
We can see that the 1st term on the RHS is a divergence(the divergence of a vector field [math]\displaystyle{ F }[/math], [math]\displaystyle{ div F = \nabla \cdot F }[/math]), and so we can apply Gauss' Divergence Theorem which states, for the region [math]\displaystyle{ R }[/math] bounded by the curve s:
- [math]\displaystyle{ \iint\limits_R \nabla \cdot F\, dA = \oint\limits_s F \cdot n\, ds }[/math]
where [math]\displaystyle{ n }[/math] is the unit vector normal to the surface.
and so,
- [math]\displaystyle{ \iint\limits_R \nabla\phi_i \cdot \nabla u\, dA = \oint\limits_s \phi_i\nabla u \cdot n\, ds - \iint\limits_R f\phi_i\, dA }[/math]
but our homogeneous boundary conditions also tell us that [math]\displaystyle{ u }[/math] is equal to zero at the boundary and so the 1st term on the RHS goes to zero:
- [math]\displaystyle{ \iint\limits_R \nabla\phi_i \cdot \nabla u\, dA = - \iint\limits_R f\phi_i\, dA }[/math]
So far, all of the operations above have been done on our unaltered, continuous differential equation. At this point, let's introduce the discritisation--and hence the approximation--that we need to make to work on a digital computer. We intend to approximate the solution [math]\displaystyle{ u(x,y) }[/math] using a linear combination of basis functions:
- [math]\displaystyle{ U(x,y) = \sum_{j=1}^m U_j\phi_j(x,y) }[/math]
The Galerkin method is being used here, as our basis functions are the same as the weighting functions used above.
- [math]\displaystyle{ \sum_{j=1}^m U_j \iint\limits_R \nabla\phi_i \cdot \nabla\phi_j \, dA = - \iint\limits_R f\phi_i\, dA }[/math]
At this point, we can introduce a symmetric 'stiffness matrix' with entries:
- [math]\displaystyle{ K_{ij} = \iint\limits_R \nabla\phi_i \cdot \nabla\phi_j \, dA }[/math]
and a vector F, with entries:
- [math]\displaystyle{ F_i = - \iint\limits_R f\phi_i\, dA }[/math]
and thus our [math]\displaystyle{ m }[/math] equations:
- [math]\displaystyle{ \sum_{j=1}^m K_{ij}U_j = F_i }[/math]
which, we see, yet again is a linear system of equations of the form Ax=b. Cue a call to a linear algebra library!
If we use linear basis functions over our domain, we will end up with the same system of linear equations to solve as we would get following the FDM:
Rather than trying to write an example program to demonstrate the FEM, it makes much more sense to look at a freely available FEM package, such as Elmer (http://www.csc.fi/english/pages/elmer). Below are images obtained by running one of the examples included with Elmer:
Other Linear Algebra Packages
Practice: PLASMA for Multicore Architectures
http://icl.cs.utk.edu/plasma/index.html
cd ../example4 make ./mxm-laplace-plasma.exe
For further examples, see e.g. INSTALL_DIR/plasma-installer_2.3.1/build/plasma_2.3.1.
Practice: MAGMA to use GPUs in Heterogeneous Systems
http://icl.cs.utk.edu/magma/index.html
Further Reading
- [1] Peter J. Olver has made available some excellent notes on the topic of solving PDEs, including a description of the FEM.